Innolitics’ DICOM Standard Browser helps users locate metadata in DICOM files, the standard radiology image format, as well as navigate the standard. It also shows users what each metadata attribute means, its type, and whether it is mandatory. Innolitics built the browser to help people learn DICOM as quickly as possible, and, by doing so, help increase compliance to the standard. Many of our users have requested that we display example DICOM values for each metadata attribute. In this article, we describe how the feature was implemented.
The process of creating this example values display was split into several different sections:

Attribute scraping 🔗
The first step was to create a script that took a single file input and output a map of all the relevant data fields and their associated values. We excluded private attributes since they are vendor-specific. Additionally, certain attributes’ value representations would not have yielded usable data—for instance, “SQ” (sequences) and “OB” (other bytes)—and these were omitted as well.
The dcmread function from the pydicom library was especially useful for extracting data from the DICOM files. For each file, we used dcmread to create a dictionary with the tags, their values, and other relevant information, such as the value representation, multiplicity, and type. Using this dictionary, it was a matter of selecting the information we wanted and placing it into a new dictionary holding only the tag and its value.
We also limited the example values’ string lengths in order to decrease the amount of data we’d eventually have to store and serve to users.
Throughout this project, we used the json library to encode our Python dictionaries as JSON. Also, the argparse library was utilized to provide users of the scripts with flexible configuration options. For this script, the ArgumentParser allowed the user to specify the input file, output file name, the maximum string length of the example values, and any tags that needed to be excluded.
Aggregation 🔗
The scraping script maps a DICOM file to a JSON file. The next step was to create an aggregation script that would combine the scraped files into a single file. We used defaultdict from the collections library to create an empty dictionary where each key maps to an empty list. For each input map, we iterated through its tags and added them to the new dictionary. If the tag already existed in the dictionary, we simply added the value to the corresponding list. All the scraped JSON objects from the previous stage were now contained in one file and we had multiple example values for each DICOM attribute.
Running the scripts on DICOM datasets 🔗
Once both the scraping and aggregation scripts had been created, it was time to run the code on as many DICOM files as we could. We downloaded publicly available DICOM directories from TCIA (The Cancer Imaging Archive). However, each DICOM directory contained potentially hundreds, if not thousands, of files. Running the scraping script on each file manually would have taken an inordinate amount of time.
To handle this, we created a bash script to run through all the DICOM directories and scrape the files within, aggregating them all at the end.

Calculating tag coverage 🔗
The initial aggregated map contained hundreds of attributes. Since the ideal end goal was to display examples for every attribute on the DICOM standard browser, we counted the number of total tags possible and compared that to the total number of DICOM tags.
A better approach to data collection 🔗

Despite running for eight hours and scraping more than 100,000 DICOM files, our initial results contained example values for only 5% of all possible tags.
This was a problem of both data acquisition and coding. While the first iteration of the bash script would have worked well for a curated and limited selection of files, it was not efficient for DICOM datasets. The issue with the files we had is that each DICOM set was organized by series and study, which often meant that much of the data within a single directory would be repeated, and thus redundant for our final JSON aggregation. Much of the data in these sets held information for a narrow group of CIODS (namely CT and MR scans). Our goal was to maximize the variety of data, not the quantity. Additionally, these subdirectories contained hundreds of files, which increased the time required for the script to run, for ultimately insignificant returns. We alleviated these issues in two steps.
First, we gathered a different and more limited group of datasets from TCIA. They were selected according to how varied their imaging modalities were, in the hopes that this would increase the tag coverage by including more CIODs.
Next, we modified the Bash script to only scrape the first file from each series. This would save time as well as reduce the size of the JSON aggregation file.
Running this new script on the curated dataset increased our tag coverage to more than 10%, doubling our previous coverage.
Displaying the results 🔗
After using a simple post-processing script to trim the number of examples for each attribute and handle duplicates, the last step was to create the React component to display the results. We added a simple React component and an associated .scss file to include the new example values in the user interface.
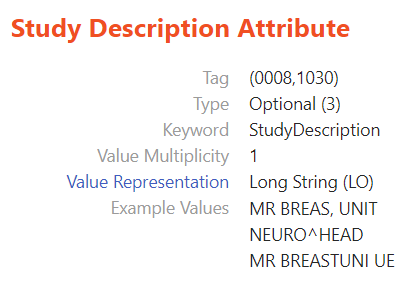
And with that, the new feature was implemented. While more examples can certainly be added in the future, we hope that the current iteration of the example data display for DICOM tags proves a useful tool for our users, and we would greatly appreciate any feedback for improvements we could make.


