Introduction 🔗

Acceptance criteria can make or break your chances of submitting your FDA application on schedule and getting clearance. Set them too high and you’ll be stuck doing more rounds of R&D. Set them too low and FDA might ask you to raise them, causing delays and risking a rejection letter. During my first 510(k) application, setting acceptance criteria took me weeks of effort. I went through many guidance documents but without any solid concrete advice. Now, over a decade and several hundred FDA interactions later, I have built an intuition around acceptance criteria that I want to share with you. It will save you some pain.
Acceptance criteria are the quantitative performance thresholds that your AI/ML medical device must meet or exceed during verification and validation testing to demonstrate that it is safe, effective, and performs as intended. They define the minimum acceptable performance before you can claim your device is ready for regulatory submission and commercial use.
You, the medical device manufacturer, are responsible for setting acceptance criteria. They represent your best-informed judgment about what performance levels will satisfy three key stakeholders: the market (for commercial viability), clinicians (for clinical utility), and regulators plus patients (for safety and effectiveness).
Think of acceptance criteria like writing requirements before sending a project to an engineering firm. Without clear specifications, the firm doesn't know when they're done and they could keep refining indefinitely. Similarly, your R&D team needs defined performance thresholds to know when testing is complete. Without acceptance criteria, development can continue with unbounded time and resources, never reaching a clear stopping point for validation.
This guide is highly opinionated. It incorporates learning from over a decade of specialized AI/ML SaMD engineering and getting devices cleared through FDA rigor. Your mileage may vary. While these rules of thumb work most of the time, each device is different and please consult a qualified consultant before you run with the recommendations in this article.
Additionally, AI and Python scripts processed thousands of 510(k) summaries to extract the acceptance criteria used in the plots and recommendations. While efforts were made to spot-check and validate the results, some errors may have slipped through. That said, these findings align with my intuition built over years in the industry, so I'm confident the overall recommendations are sound.
About the Author 🔗

Yujan Shrestha, MD is a physician–engineer and Partner at Innolitics specializing in AI/ML SaMD. He unites clinical‑evidence design with software and regulatory execution. He uses his clinical, regulatory, and technical expertise to discover the least burdensome approach to get your AI/ML SaMD to market as quickly as possible without cutting corners on safety and efficacy. Coupled with his execution team, this results in a submission‑ready file in 3 months.
Why this is important for our 3 month FDA submission guarantee 🔗

Early in my career, I spent weeks debating which approach to take for setting acceptance criteria. Looking back, this decision should have taken only a few hours. The problem was simple: concrete guidelines didn't exist. FDA guidance documents, standards, and regulatory templates spoke in abstractions and broad generalities—none offered specific numerical starting points or practical examples for real-world device development.
Hitting our 3-month 510(k) submission guarantee becomes nearly impossible if you spend an entire month deciding on acceptance criteria. Instead, I'll clarify the confusing topics and smooth out the roadblocks I've encountered.
Executive Summary 🔗

Before you set your acceptance criteria, you need to first identify the device outputs to even set criteria for. These device outputs can be categorical scores (e.g. low/medium/high or positive/negative), continuous (e.g. HU values, millimeters, seconds, etc), or segmentations (e.g. cardiac plaque ROIs, liver segmentations, lung nodule bounding boxes, etc).
For each device output:
- Pick a statistical analysis methodology
- For categorical, pick Sensitivity/Specificity for binary or weighted Kappa for multi-class
- If you can easily change your algorithms operating point, I recommend including AUC too.
- For segmentation outputs, pick Dice or HD95
- For continuous outputs, pick Bland Altman limits of agreement
- You can pick other statistical analysis methods but these are my favorites and is supported by the frequency seen in 510(k) summaries backed up in the analysis in this article.
- For categorical, pick Sensitivity/Specificity for binary or weighted Kappa for multi-class
- Set your acceptance criteria
- Pick a reasonable guess based on the rules of thumb:
- AUC of 0.9. Sensitivity/Specificity of 0.8. Kappa of 0.85. Dice of 0.8. Bland Altman LOA and Bias +/-10% and +/-2% of expected mean physiological values respectively.
- You can go lower but the above are what I consider reasonable starting points and are backed up by the analysis in this article
- I find significantly easier to start from an initial guess and refine later as you gather more evidence.
- Pick a reasonable guess based on the rules of thumb:
- Justify your acceptance criteria by finding evidence from one or more anchors in order of highest strength to lowest:
- Special controls are the strongest, predicate device 510(k) summary, reference devices, clinical observations in literature, and expert opinion being the weakest
- If the anchor only contains observed values, you can use a rule of thumb to estimate the acceptance criteria
- For Dice, sensitivity, and specificity subtract 0.1 and round to the nearest 0.05.
- For AUC and Kappa subtract 0.05 and round to the nearest 0.05.
- For Bland Altman Bias and LOA, multiply the point estimates by 125%
- The above estimates are backed up by the analysis in this article
Materials and Methods 🔗

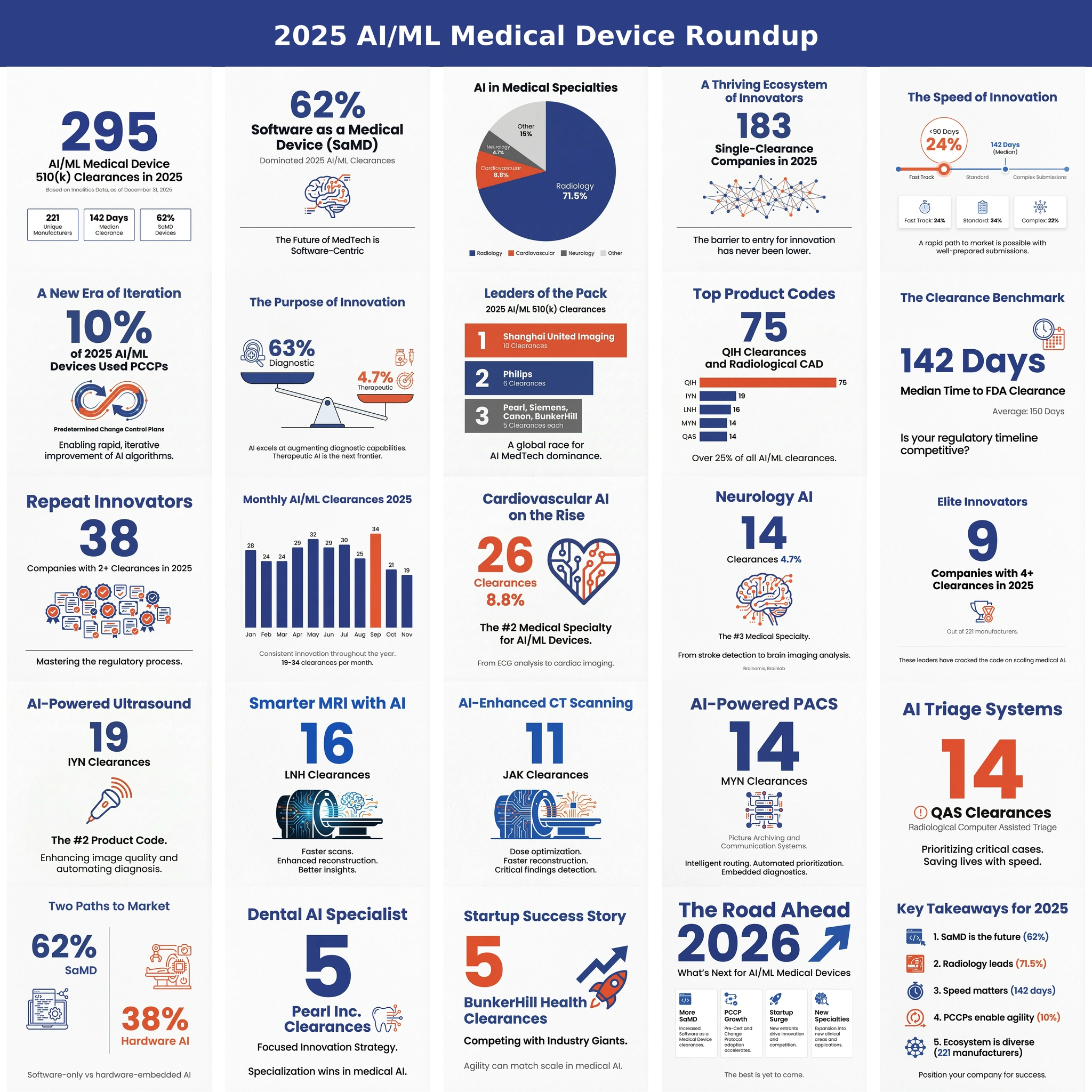
Starting with 97k+ publicly available 510(k) and De Novo PDFs from FDA’s website, the documents were OCR-processed and converted to markdown for easy LLM processing. From these, 784 submissions were identified as AI/ML SaMD using the Innolitics 510(k) Browser. Of these, 673 summaries included at least one quantitative performance metric or acceptance criterion. LLM prompts and post-processing scripts were used parsing yielded 2,217 acceptance-criteria instances and 6,396 observed-value entries, which were then aggregated and visualized using Python using the Cursor IDE, claude-4.5-sonnet LLM, and manual code review. A random sampling of extracted values were manually checked. All results were double checked to match my intuition.
Each analysis methodology (e.g. Dice, Sensitivity, Specificity, AUC, etc) will have the following plots generated:
- Histogram of acceptance criteria values
- Histogram of observed values
- Histogram of observed - acceptance criteria values. Only observed values with matching acceptance criteria are used for this. Useful to get a sense of how acceptance criteria and observed values are related.
- Confusion matrix like visualization of observed vs acceptance criteria. Only observed values with matching acceptance criteria are used for this. Useful to get a sense of how acceptance criteria and observed values are related.
Note: Case level Dice scores are typically aggregated together using a mean, median, or other methods. However, I am lumping them all together this anlaysis. I do not think this will negatively impact the takeaways.
For sensitivity and specific, an additional set of confusion matrix like plots are shown where there are paired sensitivity and specificity are available
- Confusion matrix like visualization of paired sensitivity and specificity acceptance criteria
- Confusion matrix like visualization of paired sensitivity and specificity observed values
Additionally, some aggregate plots were created:
- Summary of key acceptance criteria metrics to summarize acceptance, observed, and difference.
- Product code frequency histogram
- Metric type frequency histogram
- CAD category type frequency histogram (CADq, CADe, CADx, CADt, CADa/o)
- Number of acceptance criteria / observed values per devic
Results 🔗
Product Code and CAD Type Analysis 🔗


Most acceptance criteria analyzed came from quantitative imaging devices (CADq), followed by computer aided detection, triage, then diagnosis. A large number of acceptance criteria and observed values come from the QKB and QIH product code. Of note, the QKB product code (Radiological Image Processing Software For Radiation Therapy) often contains tens to hundreds of segmentation targets and derived quantitative imaging values. While I do not believe the increased contribution of QKB skews the results adversely, it is still worth pointing out.
Number of Criteria per Device 🔗


Most analyzed devices contributed about two acceptance criteria. However, some devices (especially in the radiation oncology space) contributed over 100. This is reflected in the large number of QKB product code and large frequency of Dice scores observed. I do not feel this impacts the recommendations provided by this article.
Overall Metric Types 🔗

Note that we have a sufficient representation of Dice, Sensitivity, Specificity, HD95, Kappa, AUC, Time to Notification, and others but other metrics like false positives per image, wAFROC, and false positives per image (FPPI) require more analysis. These will be analyzed in a subsequent article.
Analysis 🔗
Summary 🔗
Below is a summary of the metrics analyzed in this article.

| Metric | Median Observed | Median Acceptance | Median Paired Difference (Observed − Acceptance) |
|---|---|---|---|
| Dice | 0.860 | 0.690 | 0.130 |
| Sensitivity | 0.896 | 0.800 | 0.113 |
| Specificity | 0.925 | 0.800 | 0.114 |
| AUC | 0.957 | 0.950 | 0.030 |
| Kappa | 0.862 | 0.850 | 0.048 |
| ICC | 0.920 | 0.800 | 0.080 |
| HD95 (mm) | 2.300 | 4.770 | -1.870 |
| TimeToNotification (s) | 46.835 | 210.000 | -59.300 |
Dice Score 🔗
Even though devices typically report mean or median Dice scores, I have lumped together all Dice scores into one category for simplicity. From the below analysis, we can make a few key observations:
- Observed Dice score is reported more often than Acceptance Dice but not by as much as other metrics
- Observed Dice is on average 0.145 higher than the matching acceptance criteria Dice
- The median observed Dice score is 0.86
- The median Dice acceptance criteria is 0.69 but there appear to be two clusters around 0.8 and 0.65 that warrants further investigation.
- This analysis ignores the size of the target. Smaller targets have lower acceptance criteria than larger ones.




Sensitivity 🔗
From the below analysis, we can make a few key observations:
- Observed Sensitivity is reported much more often than Acceptance criteria
- Observed Sensitivity is on average 0.1 higher than the matching acceptance criteria
- The median observed Sensitivity is 0.88
- The median Sensitivity acceptance criteria is 0.8




Specificity 🔗
From the below analysis, we can make a few key observations:
- Observed Specificity is reported much more often than Acceptance criteria
- Observed Specificity is on average 0.1 higher than the matching acceptance criteria
- The median observed Specificity is 0.92
- The median Specificity acceptance criteria is 0.8
- As expected, these numbers are pretty close to Sensitivity




Paired Sensitivity and Specificity 🔗
Sensitivity and specificity alone are not that useful. Even a test that always outputs positive results is 100% sensitive. Therefore, I did an analysis of paired specificity / sensitivity values to make sure the observations are consistent with the previously discussed analysis of the two independent from one another.
From the below analysis, we can make a few key observations:
- The acceptance criteria is strongly 0.8 and 0.8 for sensitivity and specificity respectively
- The observed sensitivity and specificity cluster around 0.9 and 0.95 respectively
- I noticed only 379 paired observations were found whereas ~1000 unpaired observations are found. This likely means the algorithm I used to find the paired values was unable to match all of them. A subsequent analysis using a targeted LLM query will likely yield a better analysis.


HD95 🔗
The following observations can be made:
- More observed than acceptance values are seen
- The average acceptance value is 5.5mm
- The average observed value is 3.3mm
- On average, the acceptance value is 2.86mm larger than the observed value
- This analysis ignores the size of the target. Smaller targets could have tighter acceptance criteria than larger ones and vice versa for larger targets.




Cohen’s Kappa 🔗
From the below analysis, we can make a few key observations:
- Observed Cohen’s Kappa is reported much more often than Acceptance criteria
- Observed Cohen’s Kappa is at median 0.05 higher than the matching acceptance criteria
- The median observed Cohen’s Kappa is 0.86
- The median Cohen’s Kappa acceptance criteria is 0.85
- Note the sample size is quite low so further analysis is needed on a case by case basis in a future article




Bland Altman Bias vs Limits of Agreement 🔗
Comparing Bland Altman Bias and Limits of Agreement (LOA) between devices usually does not make sense because it is difficult to compare apples to apples exactly. Instead, I reviewed a couple of cases to get a qualitative sense of how much LOA and Bias relate to the mean value of the physiological signal being measured. I feel this ratio is a better way to generalize the findings to new scenarios.
Here are some key observations:
- These analysis is limited by sample size. A future analysis will look at Bland Altman specifically. A specific prompt for Bland Altman will likely yield better results compared to the generic prompt used for this article.
- Qualitatively, setting bias acceptance criteria to +/- 2% of the mean physiological value appears justified
- Qualitatively, setting LoA to +/- 10% of the mean physiological value appears justified
| K# | Metric | Mean physiological value (estimated) | Observed BA LoA | Mean physio value to LoA ratio | Bias (mean diff) | Bias ratio |
|---|---|---|---|---|---|---|
| K232613 | Cardiothoracic ratio | 0.460 (unitless) | [-0.033, 0.035] | 0.0761 (7.61%) | +0.001 | 0.0022 (0.22%) |
| K232613 | Heart:Chest area ratio (proxy μ uses CTR) | 0.460 (unitless) | [-0.028, 0.035] | 0.0761 (7.61%) | +0.004 | 0.0076 (0.76%) |
| K233080 | Liver attenuation | 60.000 HU | [-7.800, 7.000] HU | 0.1300 (13.00%) | −0.400 HU | 0.0067 (0.67%) |
| K251455 | EF vs manual contrast | 60.000 %-points | [-7.830, 9.260] | 0.1543 (15.43%) | +0.715 | 0.0119 (1.19%) |
| K251455 | EF vs manual tracing | 60.000 %-points | [-9.010, 11.450] | 0.1908 (19.08%) | +1.220 | 0.0203 (2.03%) |
| K250045 | Aortic root max diameter | 32.000 mm | [-0.640, 0.630] mm | 0.0200 (2.00%) | −0.005 mm | 0.0002 (0.02%) |
| K250045 | Aortic root min diameter | 32.000 mm | [-0.560, 0.560] mm | 0.0175 (1.75%) | +0.000 mm | 0.0000 (0.00%) |
| K250045 | Aortic root perimeter (μ from 32 mm) | 100.531 mm | [-2.200, 2.600] mm | 0.0259 (2.59%) | +0.200 mm | 0.0020 (0.20%) |
| K250045 | Aortic root area (μ from 32 mm) | 804.248 mm² | [-15.500, 19.000] mm² | 0.0236 (2.36%) | +1.750 mm² | 0.0022 (0.22%) |
| K250040 | EDV | 91.000 mL | [-9.610, 11.500] mL | 0.1264 (12.64%) | +0.945 mL | 0.0104 (1.04%) |
| K250040 | ESV | 34.500 mL | [-5.610, 3.800] mL | 0.1626 (16.26%) | −0.905 mL | 0.0262 (2.62%) |
| K250040 | EF | 60.000 %-points | [-3.020, 4.680] | 0.0780 (7.80%) | +0.830 | 0.0138 (1.38%) |
| K171245 | Tissue oxygenation (StO₂) | 80.000 % | [-8.563, 8.596] % | 0.1075 (10.75%) | +0.016 % | 0.0002 (0.02%) |
| K151940 | Respiratory rate | 16.000 brpm | [-0.720, 0.750] brpm | 0.0469 (4.69%) | +0.015 brpm | 0.0009 (0.09%) |
| K150754 | Axial length | 24.000 mm | [-0.070, 0.050] mm | 0.0029 (0.29%) | −0.010 mm | 0.0004 (0.04%) |
AUC 🔗
I make the following observations:
- The mean observed AUC is 0.93
- The mean acceptance criteria is 0.89
- On average, the observed AUC is 0.048 higher than the matching acceptance criteria




FAQ 🔗
Conclusion 🔗
Our analysis across hundreds of FDA submissions reveals patterns in acceptance criteria and observed values culminating in a formulaic way to set acceptance criteria.
These analysis, derived from real-world clearances, provide a data-driven foundation for setting your own acceptance criteria. While your specific thresholds should be justified by clinical context, predicate devices, and literature, these patterns offer a practical starting point that has proven acceptable to FDA reviewers across diverse AI/ML applications.
If you have a working product and want to get to market ASAP, reach out today. Let’s schedule the gap assessment, finalize claims and thresholds, and put a date on your submission calendar now. We can add certainty and speed to your FDA journey. Our 3‑Month 510(k) Submission program guarantees FDA submission in 3 months with clearance in 3 to 6 months afterwards. We are able to offer this accelerated service because, unlike other firms, we have physicians, engineers, and regulatory consultants all in house with the focus of AI/ML SaMD. We leverage our decades of combined experience, fine tuned templates, and custom built submission software to offer a done-for-you hands-off turnkey fast 510(k) submission.