Introduction 🔗
Background 🔗
If you're reading this article, you likely already understand the utility of large language models, audio models, video models, and other similar technologies. Your main concern might be whether your application qualifies as a medical device and how to get it cleared by FDA. Currently, no one has achieved FDA clearance for such models, although they are used in clinical practice under general wellness or clinical decision support guidelines. The information presented here is my expert opinion on how to obtain FDA clearance or approval for these models, based on 12 years of experience in the medical device industry and a pre-submission meeting with FDA that confirmed my understanding of how FDA views machine learning and large models.
Understanding FDA Regulation 🔗
In my opinion, most text-based algorithms, using large language models, are not currently enforced by FDA because they either fall into a general wellness category or a non-device clinical decision support category. However, any application involving audio, video, images, or signal processing is considered a medical device and is subject to regulation. Given the rapid proliferation of foundation models and the increased associated risks, I believe FDA will soon start regulating even text-based algorithms in response to adverse events.
Preparing for Future Regulations 🔗
Even if your application is not currently under FDA enforcement discretion, it is wise to prepare for potential regulation. Being proactive will give you a competitive edge when regulations tighten, and competitors may be forced to withdraw their products from the market. Preparing now ensures that you are ahead of the curve and ready to comply with future regulatory requirements.
What is in this article? 🔗
In this article, I will discuss best practices and key tips to maximize your chances of getting your foundation model-based medical device FDA approved. I'll also share a real FDA pre-submission for a large language model. Finally, I'll cover some frequently asked questions and outline qualities to look for in a regulatory consultant who can effectively explain such cutting-edge technology to FDA.
Best Practices 🔗
To achieve FDA clearance for a medical device, manufacturers must meet several key requirements. First, you need to demonstrate that your device is effective—meaning it performs as claimed. Second, you must prove that it is safe, ensuring it doesn't pose unacceptable risks to patients. Third, you need to show that the underlying technologies are sound, that your results aren't accidental, and that the device can generalize beyond your test dataset without undiscovered dangerous failure modes.
Like a court case, absolute certainty isn't required, but you must prove your case beyond a reasonable doubt. Here’s a step-by-step guide on how to justify to FDA that you have met these requirements:
Addressing Large Model Challenges 🔗
The primary issue with large models is their hidden failure modes and the lack of extensive real-world usage where these failures can be identified and analyzed. While cybersecurity aspects are beyond this discussion, clinical justification is crucial. Here are some best practices:
- Use More Deterministic Models: While complete determinism may not be possible, you can tune parameters like the starting seed and temperature to make the model as deterministic as possible. Showing best efforts in this area is important.
- Check Your Test Data Set Prospectively: Ensure your test dataset includes data that hasn't been used to train the foundation model to avoid data contamination. Foundation models are trained on diverse datasets, which can inadvertently include your test data. Maintain standard best practices, such as using multi-institutional data, diverse patient demographics, and including challenging, rare cases.
- Freeze Model Weights: Prefer models where you can freeze the weights to prevent changes. FDA prefers software implementations with strict version control of all dependencies, ensuring the same configuration is used from testing to production. Open source models like Meta’s Llama 3.1 might be more suitable for this requirement compared to some cutting-edge API services.
- Start Simple and Low-Risk: Begin with low-risk applications before advancing to higher-risk ones. For instance, instead of replacing a diagnostic test, start by creating a screening test for it. This allows you to fall back on the diagnostic test as a risk control measure, reducing the risk of false positives from your device. Conservative approaches are more likely to succeed with FDA.
-
Add More Data When in Doubt: The larger and more uncertain the model, the greater the burden of proof. Here are my recommendations for sample sizes based on the type of AI software:
- Traditional segmentation algorithm: 100-200 test samples.
- Traditional classification algorithm: 200-500 test samples.
- Classification/quantification algorithms relying on segmentation: 100-200 test samples.
- Large audio models: 1,000-50,000 test samples, depending on risk and explainability.
For example, claiming to diagnose diabetes from skin temperature readings requires more evidence due to its lower scientific feasibility. In contrast, diagnosing suspected COVID from a cough sample requires less evidence due to stronger scientific support.
Making Your Case to FDA 🔗
Convincing FDA is similar to a negotiation, where both parties need to give and take to reach a consensus. No quantitative formula guarantees success, which is why a knowledgeable consultant with expertise in machine learning, data science, software engineering, and a medical background is invaluable. They can help argue your case effectively, balancing the various considerations and requirements. An expert negotiator identifies what is important to both parties and what is easiest for both to perform, maximizing the value-to-cost ratio on both sides, and resulting in a win-win situation.
For instance, FDA places great importance on having your software configuration tightly controlled. This is relatively easy to achieve if the software is designed appropriately. Therefore, I advise you to address FDA's concerns in this area and use the goodwill gained to negotiate on more burdensome requirements, such as repeating your entire validation study.
LLM Presubmission Example 🔗
Anyone trying to get a large model through FDA right now should, in my opinion, pursue a pre-submission (pre-sub) or a breakthrough device designation. When Llama 2 was released, my team and I conducted a pre-sub to gather FDA's impressions of this new technology. We chose to wait for Llama 2 because it was the first large language model that was open-source, could be run locally with locked weights, and was relatively capable for basic tasks.
In our case, we aimed to use a large language model to analyze the DICOM series description to determine if contrast was present in the image. The results were very impressive, likely due to the model's foundational training, which included medical texts. For example, it could deduce that a CTA image was likely to contain iodinated contrast based on its foundational understanding of common CTA protocols in radiology.
If you are unfamiliar with the steps involving a pre-sub, here they are.
Step 1: Send your Presub to FDA. Wait 60 days for a response.
Step 2: FDA responds 1 week before the agreed upon meeting date.
Step 3: Send your presubmission presentation to FDA ahead of time at least 48 hours.
Step 4: Send FDA your meeting minutes
Step 5: FDA sends back their revised minutes and it becomes part of the official record.
FDA LLM Presubmission Request 🔗
We submitted the following pre-sub to FDA, incorporating both a convolutional neural network (CNN) and a large language model (LLM). We chose to include both models because relying solely on the LLM might lead to it being considered not a medical device. Additionally, the CNN provides an extra layer of risk control in case the LLM is unable to perform adequately or performs poorly.
Indications for Use 🔗
Contrast Llama is a software device intended for detecting contrast agent in CT scans and the body region scanned for the purpose of external networking, communication, processing and enhancement of CT images in DICOM format regardless of the manufacturer of CT scanner or model.
Contrast Llama may be run stand alone or as part of another medical device.
Device Description 🔗
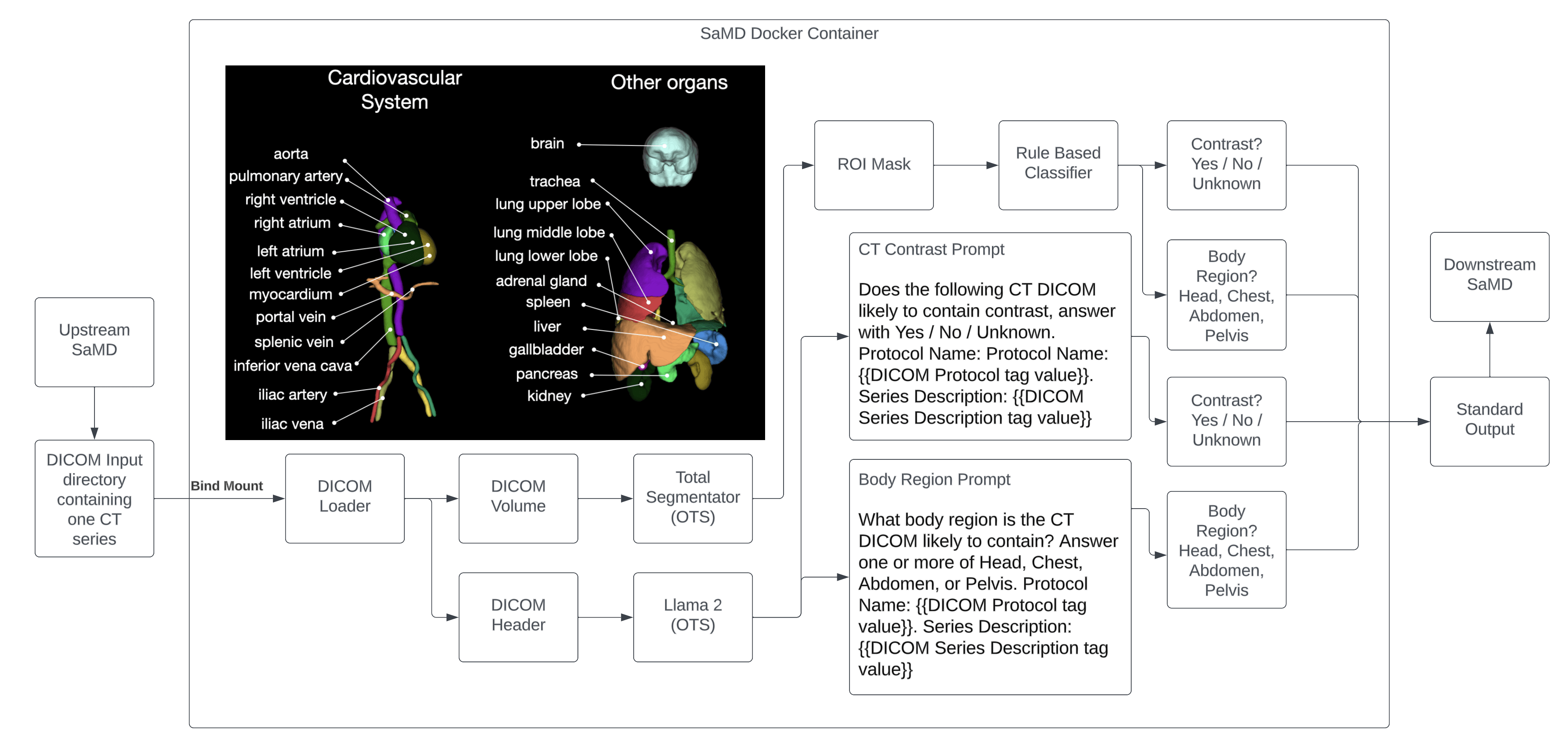
The device uses a large language model (LLM) and convolutional neural network (CNN) to determine the following characteristics about the input CT DICOM Series.
- Presence or absence of contrast agent present in the CT scan
- The body region scanned (Head, Chest, Abdomen, and/or Pelvis)
In order to perform the above, two key off the shelf software libraries are used: Total Segmentator and Llama 2.
The Total Segmentator is an OTS library that segments 104 different anatomical structures in a CT scan. Of these, 4 are used to achieve the device’s intended use: Brain, Lung, Heart, Liver, Pelvis, and Bladder.
The Llama 2 library is an OTS large language model library that is trained on a large corpus of text data. The following prompts are used, in a zero shot manner, to perform the device’s intended use:
Contrast prompt:
Does the following CT DICOM likely to contain contrast, answer with Yes / No / Unknown. Protocol Name: Protocol Name: {{DICOM Protocol tag value}}. Series Description: {{DICOM Series Description tag value}}
Example:
Input: 3D Reconstruction - Bolus Tracking
LLM Output: The description mentions "3D reconstruction" and "bolus tracking". These are features commonly used in CT imaging to reconstruct detailed 3D images of organs and tissues. The likelihood is high that the series will contain contrast, so the answer would be Yes.
Body region prompt:
What body region is the CT DICOM likely to contain? Answer one or more of Head, Chest, Abdomen, or Pelvis. Protocol Name: {{DICOM Protocol tag value}}. Series Description: {{DICOM Series Description tag value}}
Example:
Input: ProtocolName (0018,1030) 5.52 BURST 25-30 BMI TAVR PROTOCOL (NO P3T) 75-113BPM. SeriesDescription (0008,103e) ECG+BURST 75% EDITED
LLM Output: Based on the information provided in the DICOM header, the body region that is likely to contain the CT image is the Thorax (Chest). This is indicated by the ProtocolName (0018,1030) value of "BURST 25-30 BMI TAVR PROTOCOL (NO P3T)" which suggests that the CT image was acquired as part of a thoracic aortic valve replacement (TAVR) procedure. Additionally, the SeriesDescription (0008,103e) value of "ECG+BURST 75% EDITED" further supports the interpretation that the image is related to the chest region.
Llama 2 OTS is downloaded and all inference is run locally on OTS hardware without any requirement to communicate to the internet. The weights are frozen (non-adaptive) and the temperature parameter is set to 0 to enhance determinism. Test cases are run multiple times to ensure the outputs of the Llama 2 OTS are reproducible and repeatable.
Similarly, the Total Segmentator OTS library is downloaded and all inference is run locally on OTS hardware without any requirement to communicate to the internet. The weights are frozen (non-adaptive) and the outputs are deterministic. Test cases are run multiple times to ensure the outputs of Total Segmentator are reproducible and repeatable.
Neither Total Segmentator or Llama 2 are trained or fine tuned by the medical device manufacturer. Total Segmentator or Llama 2 have not been trained on the Validation Ground Truth dataset.
A Docker image fully encapsulating all software dependencies is the primary mechanism for software configuration management.
The outputs of Total Segmentator will be processed by a simple rule based classifier with the following rules:
- If at least 100 mL of lung is present in the CT series, the Chest is predicted to be present
- If at least 100 mL of brain is present in the CT series, the Head is predicted to be present
- If at least 100 mL of liver is present in the CT series, the Abdomen is predicted to be present
- If at least 100 mL of pelvis is present in the CT series, the Pelvis is predicted to be present
- If the mean HU of the heart, liver, kidneys, or bladder ROI is > 200, contrast is predicted to be present
The outputs of Llama 2 will be processed by a simple rule based classifier with the following rules:
- If the whole word “Head” is present in the body region prompt LLM output, the head is predicted to be present
- If the whole word “Chest” is present in the body region prompt LLM output, the chest is predicted to be present
- If the whole word “Abdomen” is present in the body region prompt LLM output, the abdomen is predicted to be present
- If the whole word “Pelvis” is present in the body region prompt LLM output, the pelvis is predicted to be present
- If the whole word “Yes” is present in the contrast prompt LLM output, contrast is predicted to be present.
Validation Ground Truth 🔗
- 200 random CT Scans are collected from 4 different institutions in the US. All CT scans are taken within the last year. The population is shown to be close to the U.S. population in terms of gender and ethnicity. About 50% of the scans have contrast, the other 50% do not. Around 25% of cases contain head, 25% contain chest, 25% contain abdomen, and 25% contain pelvis. Scans may contain multiple body regions.
- For all 200 CT scans
- A US board certified radiologist will open the CT scan in an OTS DICOM viewer and annotate the following:
- The presence or absence of contrast
- Well known anatomical landmarks are used to determine contrast presence including visual inspection of heart, liver, kidneys, and bladder.
- The phase of contrast: Arterial, Venous, or Delayed
- The mean HU value of the following structures are recorded: Heart, Liver, Kidneys, and Bladder using the following protocol:
- Pick a random point in the organ
- Measure the mean HU value using a circular ROI of 10mm in diameter
- Repeat 1-2 for 4 more random areas in the organ
- Record the mean of all 5 measurements
- The presence or absence (Boolean) of the Head, Chest, Abdomen, and Pelvis is annotated
- The Head is defined as any region with the Brain visible
- The Chest is defined as any region with the Lung visible
- The Abdomen is defined as any region above the pelvic brim and where Liver or Bowel is present.
- The Pelvis is defined as any region with the Pelvis present
- The presence or absence of contrast
- Another US board certified radiologist will review all annotations
- A US board certified radiologist will open the CT scan in an OTS DICOM viewer and annotate the following:
Performance Targets 🔗
The device is not a CADe, CADx, or CADt. The manufacturer has determined that only standalone performance testing is necessary. The performance target thresholds are determined to be clinically useful and sufficient for the risk level of the device.
- The device must reach an accuracy of >90% for all categories:
- Presence or absence of contrast based on CT DICOM volume (CNN)
- Presence or absence of contrast based on CT DICOM header (LLM)
- Presence or absence of Head based on CT DICOM volume (CNN)
- Presence or absence of Head based on CT DICOM header (LLM)
- Presence or absence of Chest based on CT DICOM volume (CNN)
- Presence or absence of Chest based on CT DICOM header (LLM)
- Presence or absence of Abdomen based on CT DICOM volume (CNN)
- Presence or absence of Abdomen based on CT DICOM header (LLM)
- Presence or absence of Pelvis based on CT DICOM volume (CNN)
- Presence or absence of Pelvis based on CT DICOM header (LLM)
- The device prediction must be within 100 HU of the ground truth at least 90% of the time for the following:
- Mean HU of heart, liver, kidneys, and bladder
Questions 🔗
- Does FDA agree the validation ground truth protocol is sufficient to prove efficacy?
- Does FDA agree the standalone performance targets is sufficient to prove efficacy?
- Does FDA agree with the configuration management strategy?
- Does FDA agree with the proposed predicate device?
FDA’s Presub Response 🔗
After 60 days of review, FDA came back with their responses. A big shout out to the agency for providing free guidance to industry at such an early stage for free.
In this pre-submission, you describe a device that uses CNN and LLM algorithms to analyze a CT scan to determine if contrast is present and which body region was scanned. However, you do not describe the clinical scenario for when/how this device is expected be used by the end user, what information is presented to the user, or how the outputs of the CNN and LLM are handled to determine the final output (i.e., contrast and body region labels). Without these details, we cannot determine the type of study that would be required for this device, or if additional validation is necessary. Additionally, given that a reader has many existing cues including order notes, patient history, DICOM information and their own interpreting abilities to determine the possible presence of contrast, we are unable to understand what benefit this device is providing or what gap it is trying to overcome. Without that understanding it is very difficult for us to understand the risks and how good is “good enough” performance, and that affects our responses to all of these questions.
We expect that clinicians are already able to know correctly if contrast was present, and if this device effectively changes a right answer to a wrong one in the clinical process it could introduce risks, and we do not understand the potential benefit.
Sponsor Question #1 🔗
Does FDA agree the validation ground truth protocol is sufficient to prove efficacy?
Official FDA Response
After review of the information provided, the Agency has concluded that the validation ground truth protocol is not sufficient to prove efficacy. See the following discussion points for further clarification:
- You propose a validation dataset of 200 random CT scans from 4 different US institutions that is to represent the US population with regards to gender and The scans will be evenly distributed by the presence of contrast (50% with and 50% without contrast) and by body region (head, chest, abdomen, and pelvis each represented as 25% of the cases) However, we do not believe 200 cases will be sufficient to validate your described Indications for Use; a device that is applicable to CT images in DICOM format “regardless of the manufacturer of CT scanner or model” (page 1). The entire US population includes children through adults and a breadth of abnormal pathologies, and there are a variety of CT scan makes, models, imaging protocols, annotation conventions (e.g., radiology reports, DICOM tag descriptors). Please provide statistical evidence and clinical justification for the sizing of the validation dataset presented in your future submission including corresponding descriptive statistics relevant subgroup analyses. Subgroup analyses do not necessarily have to be statistically powered. This information will be used to assess bias, generalizability, and the relevant population for your device. While your device may or may not be a CADe algorithm, we recommend you refer to FDA’s guidance on evaluation of CADe systems (Computer-Assisted Detection Devices Applied to Radiology Images and Radiology Device Data - Premarket Notification [510(k)] Submissions | FDA) for more details and general information about types and levels of detail we would like to see in order to understand and assess the design and performance of a machine learning algorithm.
- In your submission, please consider the following patient demographics: age, sex, race, and ethnicity; and provide supporting evidence that your dataset matches the US population. In addition, please provide information on the source of your dataset and the names/locations of selected sites.
- Your device uses Total Segmentator to segment specific body organs and extract the organ’s volume. The volumes (brain, heart, lungs, liver, kidney, bladder, pelvis) are used to determine contrast presence and body Please also include a distribution of the relevant pathologies for these organs, CT scan makes and models, acquisition protocols, and reconstruction methods present in your final dataset. This will be used to assess device generalizability.
- You describe that Llama 2 will use the DICOM tags “Protocol Name” and “Series Description” as inputs in the contrast and body region prompts. Please provide descriptive statistics of the breadth of protocols assessed in your validation dataset. This will be used to assess device
- It is unclear what problem you are trying to address. Your validation protocol (pages 4-5) describes the use of two US board certified radiologists to determine the reference standard. One radiologist collects the reference standard annotations, and then, a second radiologist reviews them. From your protocol description, the radiologist will not know whether this was a study performed with contrast or the study type (head/neck/chest/abdomen/pelvis). Instead, they will determine if contrast is present, and the body region based on anatomic landmarks. Can you please clarify why the radiologist would not have this additional information when determining the ground truth? This methodology may not be acceptable given that the explanation does not discuss the process by which you combine the multiple clinical truther’s interpretation to make an overall reference standard determination, nor how you process accounts for inconsistences between clinicians participating in the truthing process. For most interpretation tasks we are concerned with inter-reader variability affecting the reference standard. Note, it is typically recommended for the use of two readers, and a third reader in cases of discrepancy will serve as an adjudicator, however, this depends on the device output and associated risks. Due to the intended use uncertainty, no specific recommendations can be provided at this time. Please provide clarification or justification for why the radiologist is inferring information about the input image and provide an updated reference standard protocol that assesses the efficacy of the subject device in your future submission.
Sponsor Question #2 🔗
Does FDA agree the standalone performance targets is sufficient to prove efficacy?
Official FDA Response
After review of the information provided, the Agency has concluded that the standalone performance targets are not sufficient to prove efficacy. You defined the following acceptance criteria: “Device accuracy >90% for all categories” and “The device prediction must be within 100 HU of the ground truth at least 90% of the time for the following: Mean HU of heart, liver, kidneys, and bladder”. However, you did not provide the definition of the performance metrics selected, you did not elaborate on the validity of your selected reference standard, did not describe the intended use population, nor provide a description of the study population. Therefore, without this information, if your device is indicated to aid clinicians in distinguishing between the presence versus absence of iodinated contrast agents and between the presence versus absence of head, chest, abdomen, or pelvis body regions within the imaged field(s) of view, it would be premature to comment on the adequacy of your proposed success criteria. Please provide the definition of the measurements of performance metrics chosen, elaborate on the validity of your selected reference standard, clarify the intended use population, and provide a description of the study population. For additional guidance please see FDA Guidance titled *Guidance for Industry and FDA Staff - Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests. This and additional comments for each acceptance criterion proposed are provided below:
- On pages 5-6, you describe performance targets of accuracy >90% for each rule based component of the device. Without fully understanding the clinical usage of this device, we cannot determine the appropriate risk level. As such, we are concerned with a minimum performance threshold of 90%. We request additional clarification and statistical justification for this threshold In addition to the selected accuracy threshold, please provide the specificity, sensitivity, and AUROC for each category, as well as appropriate subgroup analyses, such as site, CT scanner make and model, demographics, etc. Note, accuracy is not recommended as a measurement for diagnostic performance, since accuracy = (sensitivity × prevalence) + (specificity × (1 ‐ prevalence)) and is not recommended compared to endpoints such as co‐primary sensitivity and specificity or AUC. Please also provide performance results for the segmentations and volume estimations for the relevant organs.
- On page 6, you describe a Hounsfield Unit (HU) performance target of within 100 HU for >90% of cases. We do not believe these are appropriate performance thresholds. This range of HU is large given the HU cut-off of 200 used in your rule-based logic, and you do not provide clinical or
statistical justification for the performance target. Please provide an alternate performance target plan with statistical and clinical justification.
While your device may not be a CADe algorithm, we recommend you refer to FDA’s guidance on evaluation of CADe systems (Computer-Assisted Detection Devices Applied to Radiology Images and Radiology Device Data - Premarket Notification [510(k)] Submissions | FDA) for more details and general information about types and levels of detail we would like to see in order to understand and assess the design and performance of a machine learning algorithm. Additionally, as discussed in FDA Guidance titled “Clinical Performance Assessment: Considerations for Computer-Assisted Detection Devices Applied to Radiology Images and Radiology Device Data in Premarket Notification (510(k)) Submissions”, the clinical performance assessment of the device is intended to demonstrate the clinical safety and effectiveness for its intended use, when used by the intended user and in accordance with its proposed labeling and instructions. However, based on the information provided in this submission, it is unclear how the device output affects the clinician’s interpretive process (e.g., concurrent or second read). The physical characteristics of the device output are unclear, for example it is unclear if the device identifies, marks, highlights, or in any other manner directs attention to aspects of radiology device data that may reveal abnormalities during interpretation. Based on the system architecture, the output of the device is unclear, and the risks associated with the device’s functionality are unclear.
Sponsor Question #3 🔗
Does FDA agree with the configuration management strategy?
Official FDA Response
After review of the information provided, the Agency has a number of comments regarding the configuration management strategy, or the use of an offline LLM with locked model weights, Llama 2. The following questions regarding the algorithm are provided below:
- It appears the Llama 2 output is a string/text. However, it is unclear how the prompt output is aggregated into a final output for cases with undefined or medium probability of contrast. Please clarify how the LLM contrast output creates a probability of small or unlikely. It is also unclear how the probability output is then categorized as no contrast, or unknown. It remains unclear if you will validate the performance of the LLM prompts, and if the user will have access to these outputs. Additionally, it is unclear what the potential benefit of each output (i.e., segmentator, LLM DICOM anatomical region detection, LLM DICOM contrast detection) are, or the utility of these in the clinical Therefore, without this information, the Agency cannot assess at this time the associated risks of the LLM output. Please provide clarification of the LLM output, the associated validation of the output prompt, and please clarify the device output and it’s intended use.
- In a future pre-submission, please provide an engineering description of the underlying model architecture and how it was trained. This information is required to help us understand the underlying functionality and complexity of your device and whether the safety and efficacy of your algorithm is expected to be generalizable to the different data acquisition devices or types of patients and demonstrate that your device is substantially equivalent to its cited predicate device
(21 CFR 807.100(b)(2)(ii)(B)). Specifically, we ask that you provide the following details in your future pre-submission for both models. Of note, this list also addresses details that may be only specific to Total Segmentator.
- Input data and the dimension of each input, including patient images and patient
- Pre-processing components, network type(s) and components (number of layers, activation functions, and the dimensionality of the data throughout the processing pipeline).
- Model development, including training and tuning, and model design components: transfer learning, data augmentation, regularization methods, loss functions, ensemble methods (e.g., bagging, boosting, averaging, etc.), tuning thresholds and hyperparameters, optimization methods (optimality criteria), and other documentable parameters.
- Sampling methods: Is the data set is an exhaustive set from some range of dates, a random sample, or a collection of consecutive cases?
- The distribution of the data along covariates such as patient demographics, disease conditions (e.g., positive/negative cases), and any special population subsets or enrichment.
- The distribution of the data along acquisition conditions (scanner models, important imaging parameters and protocols).
- Performance (e.g., sensitivity/specificity/ROC analysis) of your internal testing, if
- Details of any post-processing
In addition to providing this information about your algorithm, we strongly recommend you provide some information about your algorithm development in the user-facing documentation. Including information regarding algorithm development will ensure transparency to the user and the safe use of the device.
While your device may not be a CADe algorithm, we recommend you refer to FDA’s guidance on evaluation of CADe systems (Computer-Assisted Detection Devices Applied to Radiology Images and Radiology Device Data - Premarket Notification [510(k)] Submissions | FDA) for more details and general information about types and levels of detail we would like to see in order to understand and assess the design and performance of a machine learning algorithm. These documents also include additional recommendations and requirements, such as locking the device so that no further modifications are made during or after the standalone/clinical evaluations, or importance of ascertaining that the train/test datasets are completely independent of each other to avoid data leakage:
- https://www.fda.gov/regulatory-information/search-fda-guidance-documents/computerassisted- detection-devices-applied-radiology-images-and-radiology-device-data-premarket
- https://www.fda.gov/regulatory-information/search-fda-guidance-documents/clinicalperformance- assessment-considerations-computer-assisted-detection-devices-appliedradiology
Sponsor Question #4 🔗
Does FDA agree with the proposed predicate device?
Official FDA Response
After review of the information provided, the Agency is unconvinced the proposed predicate device is appropriate. You have proposed similar Indications for Use as the predicate device; however, we do not understand how the proposed device functions support your indications for use. It is unclear how the subject device “enhances the CT image” as it is stated in your Indications for Use. Although both devices employ a form of deep learning algorithm, it appears the subject device does not modify, or enhance, the CT image as described in the Indications for Use. Therefore, we cannot rely on the similarity of the Indications for Use
statements to help a comparison. Note, your indications for use should be representative of the subject device functionality, intended use environment and intended user.
Based on the information provided, it appears that your device is indicated to provide an output related to the presence versus absence of iodinated contrast agents and between the presence versus absence of head, chest, abdomen, or pelvis body regions within the imaged field(s) of view by directing attention to portions of a CT image or aspects of radiology device data. It is unclear what broader purpose this information is in aid of considering the presence of contrast should already be well documented in the medical record.
Therefore, we cannot fully understand your intended use or whether it is the same as that of the proposed predicate. Therefore, based on the limited device description provided, it appears that the device may have a new and different intended use.
Furthermore, your proposed device employs text processing technology, Large Language Model (LLM) algorithm, which may raise a new and different safety and effectiveness question, specifically, “Does the LLM algorithm correctly identify and match the contrast as annotated in the DICOM tags with the contrast detected in the image by the Segmentator algorithm?” Note, the use of large language models for contrast detection has not been previously determined to be substantially equivalent to a legally marketed predicate device within 21 CFR 892.2050.Based on the LLM functionality, it is unclear if this significant change to the design or feature of the device does not raise different questions of safety and effectiveness and that the device is as safe and effective as a legally marketed device.
To aid our further evaluation, please provide more details about the device output. The device output is described as a standard output. However, the final form of the output has not been described in totality, it is unclear how the output is used by the end user, and it is unclear how the final output is employed in the intended use environment. You should clarify how the final output of the device is affected if the output of the Segmentator and the output of Llama 2 differ in result (i.e., yes for contrast/no for contrast). Risk analysis for the combination of output scenarios should be provided to clarify to the Agency the potential safety and effectiveness risks associated with the new functionalities based on the LLM functionality. These differences may result in a new intended use if they may affect the safety and/or effectiveness of the subject device as compared to the predicate device and if the differences cannot be adequately evaluated under the comparative standard of substantial equivalence.
To demonstrate substantial equivalence to a cleared device under 21 CFR 892.2050, it may help for you to identify a device that has a similar feature or performs a similar task as the subject device. However, we are unaware of any device with a similar combination of features to what you are proposing. Due to the lack of information described above, the Agency is unable to provide further feedback regarding recommended regulatory pathway. Please consider submitting a presubmission supplement containing an enhanced device description and discussion of regulatory pathway. We also recommend submission of a draft protocol or protocol summary (including statistical analysis planning) for FDA feedback prior to collection of performance data.
For additional information, please also review relevant portions of the following guidance documents available online:
- https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/clinical‐ performanceassessment‐considerations‐computer‐assisted‐detection‐devices‐applied‐radiology
- https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/computer‐ assisteddetection‐devices‐applied‐radiology‐images‐and‐radiology‐device‐data‐premarket
- https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/software‐medical‐ devicesamd‐clinical‐evaluation
- https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/statistical‐ guidancereporting‐results‐studies‐evaluating‐diagnostic‐tests‐guidance‐industry‐and‐fda
- https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/best‐practices‐ selectingpredicate‐device‐support‐premarket‐notification‐510k‐submission
Presubmission Meeting and Response 🔗
Next, we had a meeting with FDA to discuss the concepts during an interactive one-hour meeting. Here are the minutes. Note, it is the sponsor's responsibility to take meeting minutes and give to FDA for approval.
Device Overview 🔗
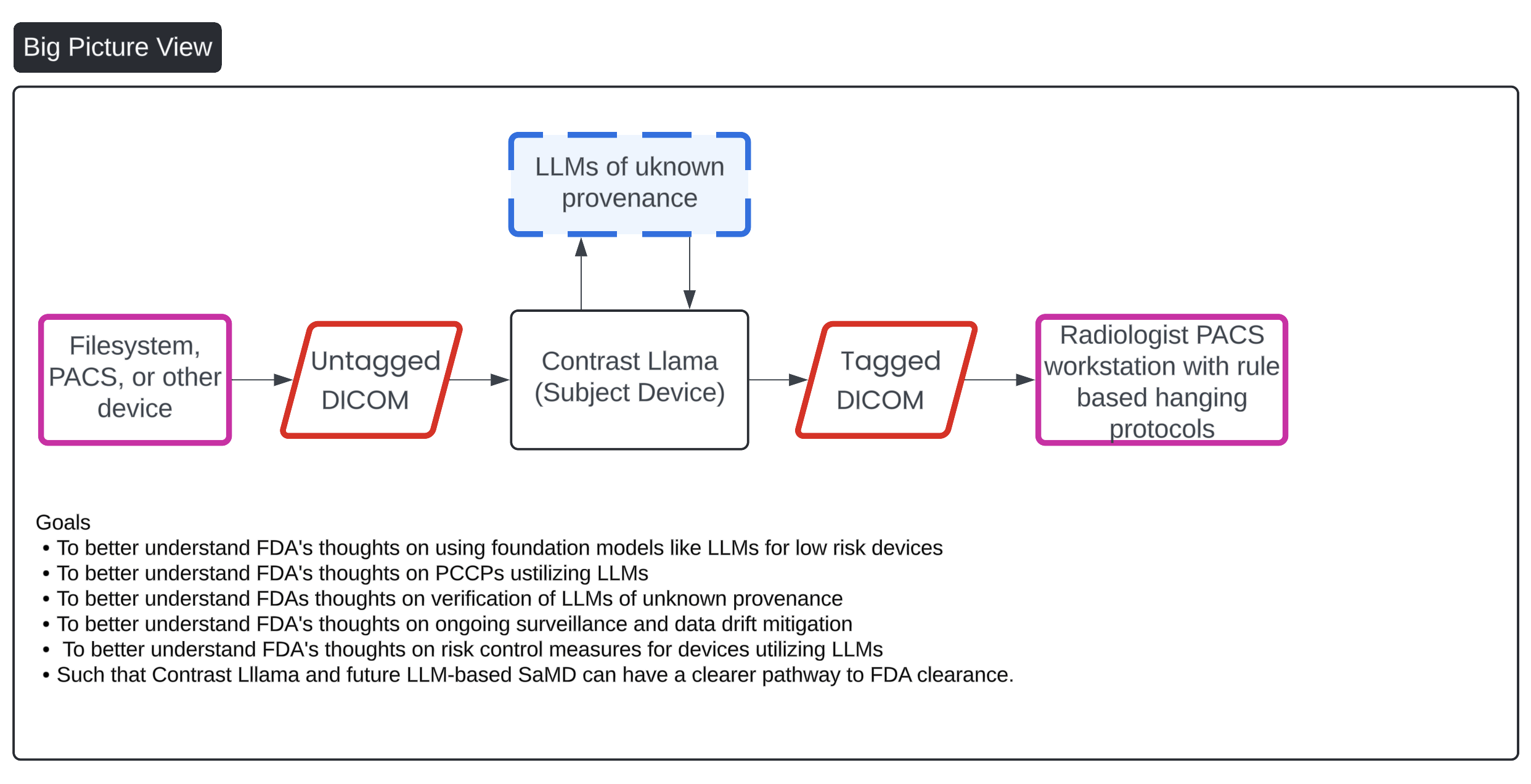
- Sponsor gave high-level overview of lnnolitics (company), and the clinical purpose for subject Specifically, sponsor stated that there are two big- picture motivations behind the subject device: 1) developing a SaMD that can be used by other SaMD manufacturers for contrast filtering and body component identification and 2) introduce LLM for use in low-risk environment while establishing best practices.
- FDA commented that there had been some confusion on the intended use of the subject device, but the Agency now has a better understanding of the motivation behind it.
- Sponsor gave an overview of the algorithm Sponsor explained that an upstream SaMD would send images to the subject device. Then, subject device would make determinations (contrastversus non-contrast and body- region identification). Subject device then sends images to a downstream SaMD. In essence, the subject device's purpose is to filter out images that downstream device is not intended to process.
- FDA had a question about the device's architecture. The Agency stated that it seems like both algorithms are working in parallel and it is not clear which output takes Sponsor stated that LLM will only be used if image data is not present. Sponsor stated that there may be a future plan to combine both algorithms if this demonstrates improved performance. However, Sponsor clarified that pilot data is needed in order to understand the device's performance. Nonetheless, the current plan is for the LLM to be used as back up.
- FDA has a question about the user's ability to see the outputs of the The Sponsor clarified that a clinician user would not see the outputs of device, as device is not intended to be used directly by this user and would be more of a blackbox. In the scenario where the device did not work as intended, this would result in no report being generated by the downstream SaMD.
Written Feedback 🔗
Abnormal Pathologies and Device Uses
- Sponsor stated that intended patient population will be adults
- Sponsor stated that dataset will be enriched with abnormal pathologies, but only with those that Sponsor believes would have an effect on the outcome of the device (e.g.,anything that would affect mean HU calculation). Sponsor believes that other pathologies like breast cancer or soft tissue cancer would not affect the outcome of the Sponsor asked the Agency if they believe this to be a valid approach for determining which pathologies should be included in data.
- FDA stated that the proposed approach, broadly speaking, should be FDA stated that it would be ideal to have a list of subgroups/pathologies for review by the Agency.
- FDA stated that it would also matter what type of downstream device the subject device is intended to be feeding
- Sponsor stated that due to the device's nature and intended use, it might not be possible to know all the types of downstream devices that the subject device could be used FDA acknowledged this and stated that Sponsor should have some sense of characterizing the device's performance for potential future use scenarios.
LLMs and Use of Synthetic Data
- FDA stated that the topic of incorporating LLM is a novel idea and more internal conversation needs to be had about these types of devices, which might affect future FDA stated that, broadly speaking, image-based Al devices should be trained in a way that will generate meaningful results.
- Sponsor asked the Agency about using synthetic Sponsor proposed having a board-certified radiologist review it. FDA stated that given their current understanding there would a tremendous amount of information needed on the model. Thus, it might be easier to provide real data.
Validation Sample Size
- Sponsor proposed using a sample size of 200 cases for testing the Sponsor stated that this sample size has been powered, but understands that the Agency has other statistical concerns besides power (e.g., generalizability). Sponsor asked the Agency for guidance in determining an appropriate sample size.
- FDA stated there is no "correct" number for sample Generally speaking, the Agency wants to ensure that there are enough cases for generalizability, e.g., different manufacturers, models, demographics and protocols that would be seen in clinical practice. FDA stated that Sponsor should provide a detailed description of the dataset, so the Agency can review it and give feedback on what is missing (e.g., other subgroups that should be considered, etc.). FDA recommended that Sponsor looks at 510(k) summaries of similar devices to understand what subgroups have been included in previous submissions. Moreover, FDA stated that the target clinical condition would also drive what is acceptable to provide (e.g., locations, confounders, etc.).
- FDA stated that depending on prevalence, consecutive collection is usually good method to However, the Agency emphasized that for LLM, FDA does not have clear criteria on what they are looking for, but welcomes Sponsor justifications.
Other LLM Concerns
- Sponsor stated that it is now possible to run LLM locally, so have reasonable understanding that it would be safe to use in FDA stated that using a "frozen/locked" model and providing a clear understanding of how it is being deployed would be recommended in future submission.
- Sponsor asked the Agency if there are any other general concerns for the use of FDA stated they haven't seen a lot of projects that use LLMs. FDA stated that a frozen application would be desirable, but if Sponsor is not able to do that, then reason to not do that then there would be a concern of testing device in a "black box" environment would be much more difficult.
- Sponsor stated that the current goal is to pick use cases that are low-risk and technological possible. FDA stated that their current expectation is to understand the details on how it was developed, layers of model, training, among other The Agency stated that they don't really know what their expectations are for LLMs, and these will be understood more as they learn more about it.
510{k) Route and Risk-Level
- FDA stated that due to the new information, the review team didn't get a chance to discuss the intended use internally, but depending on it, the 510(k) pathway may or may not be appropriate for the subject device.
- FDA stated that Sponsor needs to provide details on how the device would be considered a standalone medical
- FDA stated that Sponsor might need to may consider the Master File "program" for future third party manufacturers to use the subject device as a "component" of their own.
- FDA stated that if subject device is intended to automate something that could lead to serious harm, then this would increase the risk profile of the device. For example, using the subject device with downstream triage devices.
- Sponsor proposed clearly listing typical applications in the indications for use, but still leaving it open for general use. FDA recommended that Sponsor provides descriptions of what applications the device should be used for, what it may be used for, and what it should not be used
- FDA suggested Sponsor follows up via a 513(9g) or an additional Q-Submission. FDA stated that a 513(9g) would involve higher-level CDRH members and could provide higher-level regulatory guidance on the appropriate pathway. On the other hand, a follow up Q-Submission would dive deeper into some of the topics discussed in this meeting. The Agency stated that it is up to the Sponsor to determine which type of follow up would be preferred.
- Additionally, FDA stated that they are happy to share new guidance and expectations informally as they become available, but it is not clear when this would be possible.
Statistics of Breadth Protocols
- Sponsor asked if the Agency could share guidance on providing statistical descriptions of CT protocols.
- FDA stated that their current thinking is requesting for information like distribution of manufacturers and subgroup analysis, as these give the Agency a sense of what has been represented in the dataset. For example, if DICOM protocols will be main inputs of the device, Sponsor we should clarify the scope and breadth of the inputs. FDA stated that the spirit of the ask is to give the Agency confidence on generalizability to the intended US population. Moreover, FDA stated that they don't have specific expectations, but are learning along the way.
lnter-reader Variability
- Sponsor stated that having a triplicate read seems overly burdensome, since the tasks in question seem are relatively "basic" and thus, would not expect much reader inter-variability.
- FDA stated that they originally did not understand what device was intended for, and welcomes the Sponsor to provide a justification for inter-reader variability in future submission.
FAQs 🔗
Final Thoughts 🔗
Foundation models are at the forefront of technology right now, offering immense clinical and business potential. They can significantly reduce the data required to create clinically useful algorithms due to extensive pre-training, allowing these algorithms to generalize better than their contemporary counterparts. However, given the cutting-edge nature of foundation models, it’s important to approach their implementation with the right mindset.
Instead of asking consultants whether they have done this before—since no one has—focus on how their past experiences can prepare them to get a foundation model FDA approved. Look for consultants with the following qualities:
- Strong Data Science and Software Development Experience: This expertise allows a consultant to interpret the existing FDA guidances on traditional machine learning topics and anticipate how FDA will respond to new situations.
- Extensive Software as a Medical Device (SaMD) Experience: Many principles from SaMD apply to foundation models, with configuration management being particularly critical. Plucking low hanging fruit to buy goodwill with FDA in various areas is essential before presenting your device with cutting-edge, unproven technology.
- Clinical Experience: While not mandatory, clinical experience helps consultants understand patient risk, which is crucial since the burden of proof often revolves around risk versus benefit. This background facilitates meaningful discussions about the clinical implications of your device.
- Familiarity with Quality Systems Tooling: Especially if you haven't set up a quality system before, a consultant with experience in this area can be invaluable. Quality systems are a necessary part of any medical device entrepreneurial operating system and shapes your company’s culture for years to come.
Good luck with your foundation model. Keep dreaming and creating innovative solutions to improve patients' lives!
Revision History 🔗
| Date | Changes | |
|---|---|---|
| 2024-07-27 | Initial Version |