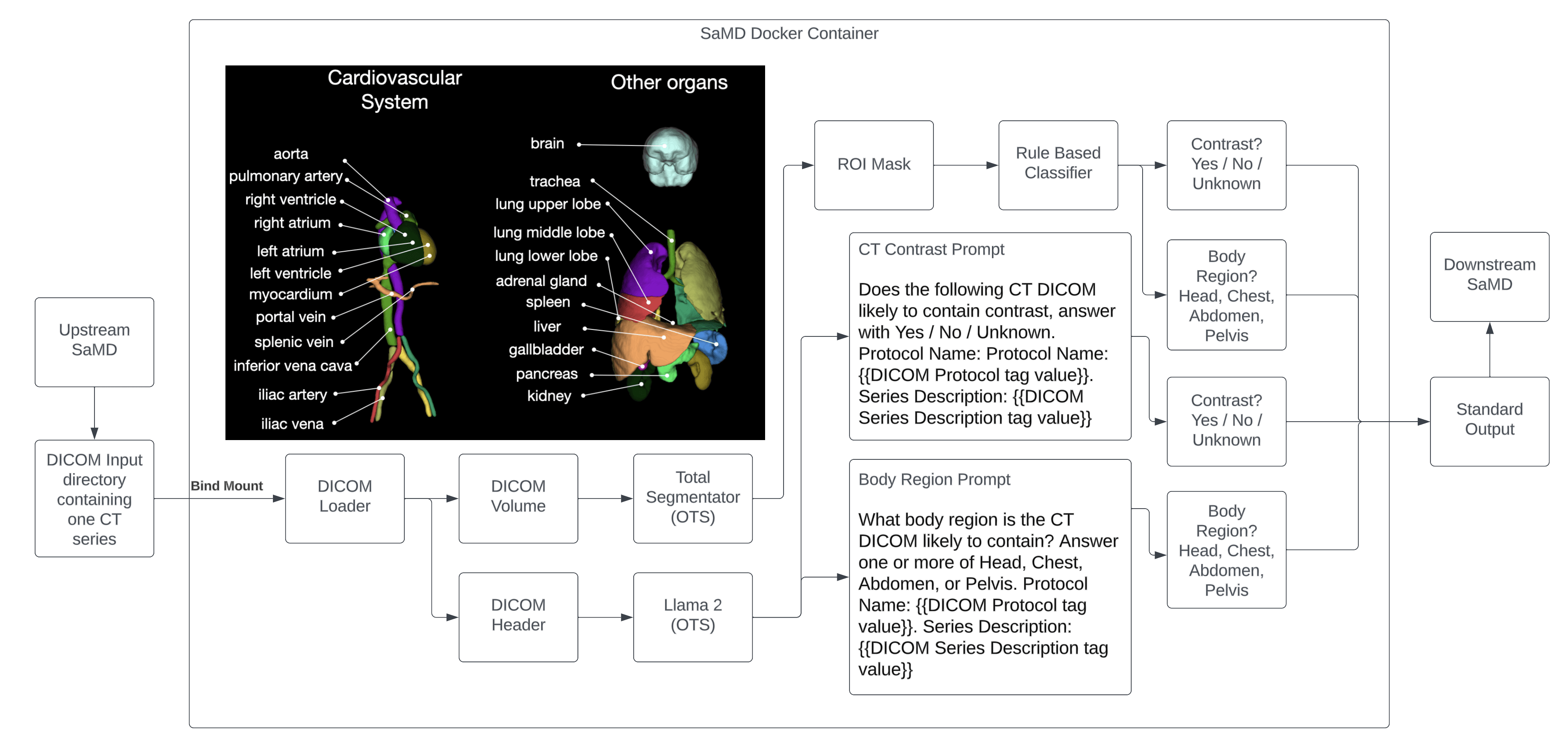
Background 🔗
There are a lot of articles online outlining the problems with foundation models and generative AI in medical devices. However, there are relatively few proposing actual solutions. While it is easy to interpolate what FDA has already released, it takes deep first-principles understanding to extrapolate into the future. Innolitics is an engineering-first consultancy that has the foundational knowledge to extrapolate into novel and unknown territory. In this thought leadership piece, we leverage our engineering-first expertise to propose potential solutions to successfully clearing medical devices with foundational models through FDA. This article is for you if you are tired of reading the sea of articles calling for regulation of foundation model applications but fail to offer concrete actionable advice.
The following is a pre-submission we have submitted to FDA with these concepts. We are eagerly awaiting feedback from FDA, but wanted to share our engineering-first strategic thinking for an open discussion.
Presub Intro 🔗
Innolitics, LLC is requesting a Q-submission supplement (S001) for Q232314 in the form of a Teleconference meeting to further continue our discussion of the Contrast Llama device. Contrast Llama is a standalone, command-line driven software medical device intended to assist healthcare professionals in the automated classification of Computed Tomography (CT) DICOM files based on their header information. The device is designed to be integrated into existing radiology workflows to enhance efficiency and consistency in image handling and interpretation processes.
This document provides additional information on key topics and highlights our specific questions for the Agency.
Additionally, a video file has been attached as part of this Q-submission preSTAR. The video includes a detailed walkthrough of the different sections found within this document. Time stamps for the video are listed below:
- 00:00: Overview (Introduction, Big Picture View Diagram, Intended Clinical Workflow)
- 02:59: Runtime Description (Runtime View Diagram)
- 09:14: LLM Based Boolean Classifier Prompt (Overview of Prompt)
- 14:05: PHI and Cybersecurity Attack Removal Prompt (Overview of Prompt)
- 15:26: Test Dataset Inventory
- 18:02: Foundation Model Training Description (Training Data Card Inventory)
- 19:11: Manual Ground Truthing Description (Description of Process, Examples)
- 23:17: Automatic Ground Truthing Description
- 24:17: Traditional ML Verification (Standalone Performance Test Plan)
- 26:57: ML Verification View for PHI and Security Threat Scrubbing (Deidentification Test Plan)
- 28:07: Non ML Verification View (Verification of Non-ML Algorithm/Components)
- 29:03: Automated ML Verification
- 34:25: Predetermined Change Control Plan
- 38:26: Conclusion
Indications for Use 🔗
Contrast Llama is a standalone, command-line driven software medical device intended to assist healthcare professionals in the automated classification of Computed Tomography (CT) DICOM files based on their header information. The device is designed to be integrated into existing radiology workflows to enhance efficiency and consistency in image handling and interpretation processes.
Specifically, Contrast Llama is intended to:
- Analyze DICOM header information from CT scans to classify images into predefined categories such as contrast-enhanced, dual-energy, body region-specific (e.g., chest, abdomen, brain), and special protocols (e.g., angiography, low-dose, pediatric, trauma).
- Provide Boolean classification outputs that can be utilized by downstream processes, including automated hanging protocols and workflow prioritization systems.
- Operate as a command-line tool, allowing for seamless integration with existing hospital information systems and PACS (Picture Archiving and Communication Systems).
- Support radiologists, technologists, and other qualified healthcare professionals in optimizing their workflow by automating the initial categorization of CT studies.
Contrast Llama is not intended for diagnostic use or to replace the professional judgment of healthcare providers. It is designed as a workflow optimization tool to be used in conjunction with, and not as a substitute for, the expertise of trained medical professionals. The device is intended for use in hospitals, imaging centers, and other healthcare facilities where CT scans are routinely performed and interpreted.
This software is to be used by trained medical professionals who are familiar with CT imaging protocols and DICOM standards. Contrast Llama is not intended for use by patients or lay persons.
Overview 🔗
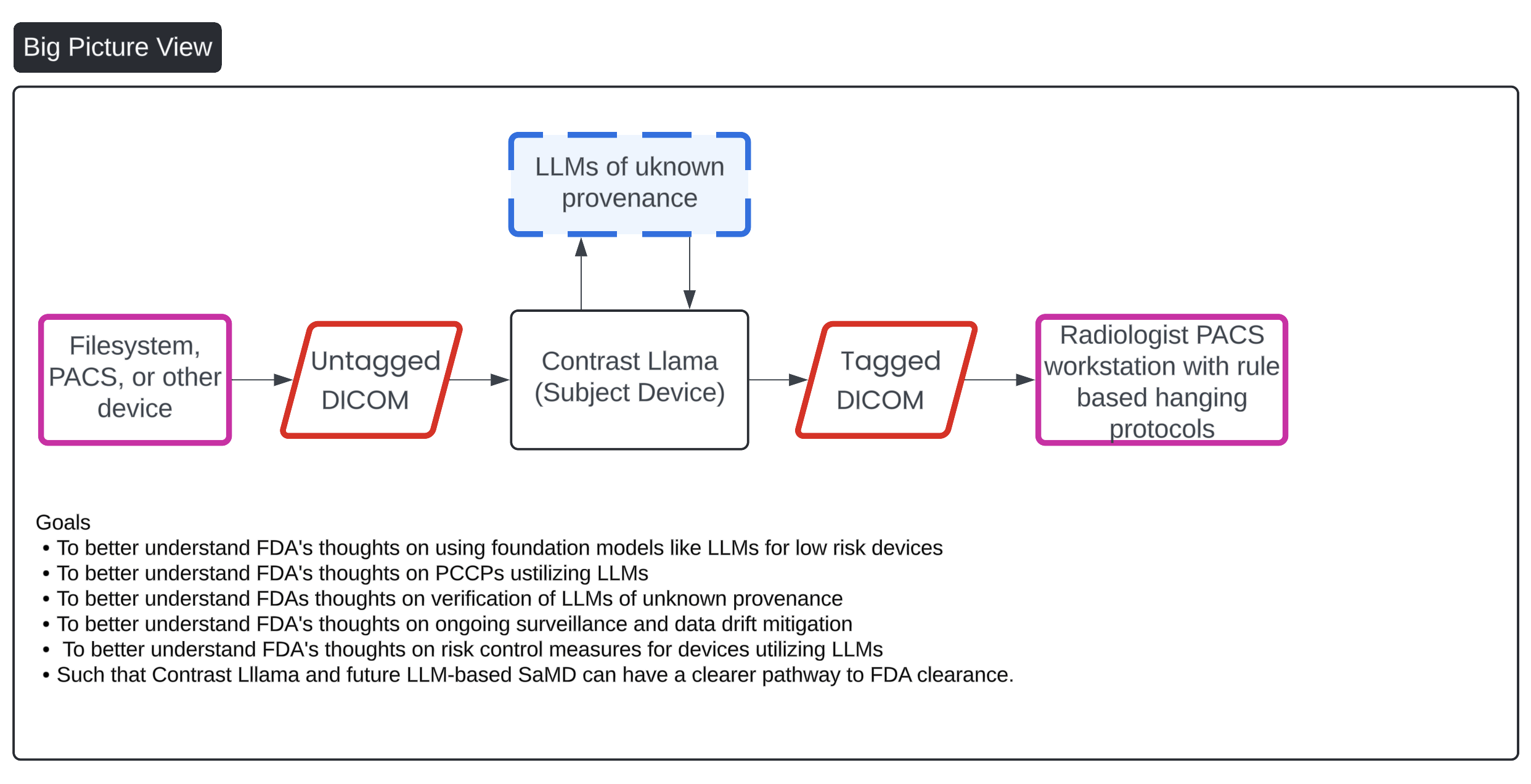
This image presents a flowchart that outlines the process of managing DICOM (Digital Imaging and Communications in Medicine) files through a system that incorporates an AI-based device called "Contrast Llama," aimed at enhancing medical imaging workflows. The process starts with untagged DICOM files sourced from external devices like file systems or PACS (Picture Archiving and Communication Systems). These untagged files are then processed by the "Contrast Llama" system, which utilizes Large Language Models (LLMs) to convert the untagged DICOMs into tagged versions, adding metadata that facilitates easier classification and downstream processing by radiologists. The final tagged DICOMs are sent to a radiologist’s PACS workstation, utilizing rule-based hanging protocols for interpretation.
Key goals listed in the chart include better understanding FDA's stance on using LLMs for low-risk devices, verification of LLMs of unknown provenance, risk control measures, and post-market surveillance of these AI-driven devices to facilitate FDA clearance for the Contrast Llama system and future LLM-based Software as a Medical Device (SaMD).
Runtime Description 🔗
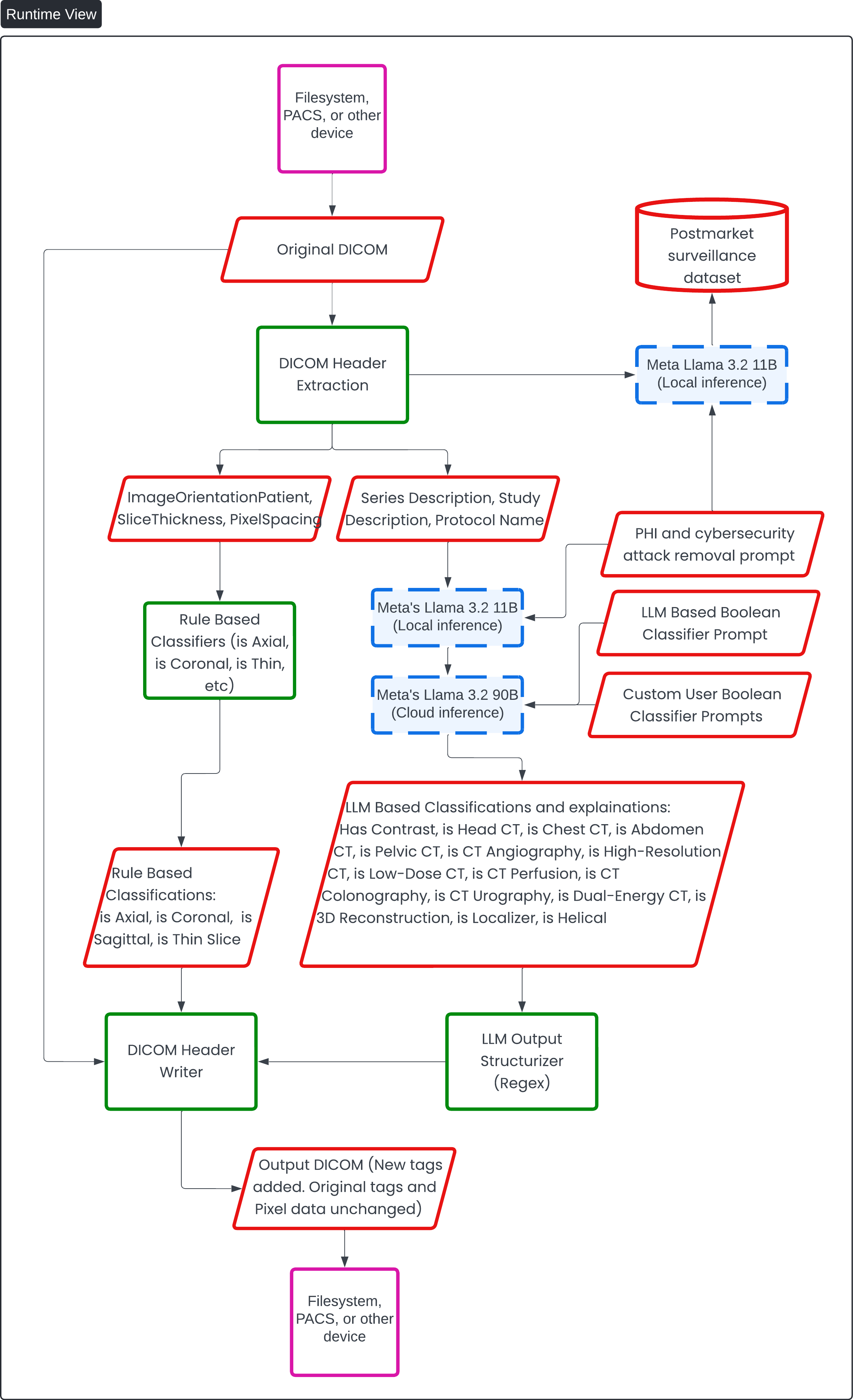
This diagram provides a "Runtime View" of the AI-driven DICOM processing system, specifically the "Contrast Llama" device. The flow of information starts with the input of DICOM files from external devices such as filesystems or PACS. The diagram then illustrates the extraction of DICOM header data, which is categorized into elements such as "ImageOrientationPatient," "SliceThickness," "PixelSpacing," and "Series Description, Study Description, Protocol Name."
The extracted header information is then divided into two streams:
- Rule-Based Classifiers: These classifiers, using traditional methods, determine attributes like whether the scan is axial, coronal, sagittal, or a thin slice. The rule-based results are fed back into the DICOM header.
- LLM-Based Classifications: Large Language Models (LLMs), including Meta's Llama 3.2 models (run locally and on the cloud), classify more complex details such as whether the image has contrast, and whether it’s a head, chest, abdomen, or pelvic CT, among others. These LLMs use both local and cloud inference. They are also responsible for PHI and cybersecurity attack removal, and custom user prompts provide additional Boolean classifier prompts for more tailored classifications.
The output from the LLMs is structured using a regex-based LLM output structurizer before the new tags are added back into the original DICOM file through a DICOM header writer. The process concludes with the output of the DICOM files, which maintain the original pixel data and tags but include the new classifications, ready to be used again by filesystems or PACS for medical imaging workflows.
Additionally, there is a post-market surveillance dataset which appears to feed into the system for continuous monitoring and updates, ensuring ongoing performance and risk mitigation.
LLM Based Boolean Classifier Prompt 🔗
Below is the prompt used by the LLM to perform the boolean classification.
You are a skilled radiologist. Your task is to analyze CT DICOM header information and identify which CT sequence classifiers are positively matched based on the data provided. You will process the DICOM headers, explain your reasoning for each match, and output a list of classifiers that are true for the given CT SERIES.
Instructions:
1. Analyze the DICOM Headers: You will be given relevant DICOM tags such as `SeriesDescription`, `StudyDescription`, and `ProtocolName`.
2. Determine the Positive Classifiers: Use the information from the DICOM headers to determine which classifiers are positively matched only to the given SERIES.
3. Explain Your Reasoning: For each positively matched classifier, provide a brief explanation based on the DICOM headers.
4. Output Format: follow the example below
5. Do Not Include Negative Matches: Do not mention classifiers that are not matched.
6. Handle Confounders Carefully: Be cautious of terms or values that might be misleading.
7. Do Not Hallucinate: Do not mention unless you are sure.
8. Spell the possible classifiers exactly as shown below.
---
List of Possible Classifiers:
1. Has Contrast: Indicates that a contrast agent was administered and used during the acquisition of this CT series, enhancing certain tissues and structures to improve image contrast and lesion detection.
2. Is Head CT: Indicates that this series is a CT scan of the head, used to assess brain structures, skull fractures, hemorrhages, and other cranial pathologies.
3. Is Chest CT: Indicates that this series is a CT scan of the chest, used to evaluate lung parenchyma, mediastinum, pleura, and chest wall for conditions such as infections, tumors, or pulmonary embolism.
4. Is Abdomen CT: Indicates that this series is a CT scan of the abdomen, used to visualize abdominal organs such as the liver, pancreas, kidneys, and to detect abnormalities like tumors, stones, or inflammation.
5. Is Pelvis CT: Indicates that this series is a CT scan of the pelvis, used to examine pelvic organs including the bladder, reproductive organs, and to detect fractures or tumors.
6. Is CT Angiography: Indicates that this series is a CT angiography (CTA) sequence, designed to visualize blood vessels by using contrast agents, useful for detecting vascular diseases like aneurysms or blockages.
7. Is High-Resolution CT: Indicates that this series uses high-resolution CT techniques, providing detailed images of lung parenchyma, useful for evaluating interstitial lung diseases.
8. Is Low-Dose CT: Indicates that this series is a low-dose CT scan, often used for lung cancer screening or in situations requiring reduced radiation exposure.
9. Is CT Perfusion: Indicates that this series involves CT perfusion imaging techniques to evaluate blood flow through tissues, aiding in the assessment of ischemia or infarction.
10. Is CT Colonography: Indicates that this series is a CT colonography, also known as virtual colonoscopy, used to screen for polyps or colorectal cancer.
11. Is CT Urography: Indicates that this series is a CT urography, specialized for imaging the urinary tract including kidneys, ureters, and bladder, often using contrast.
12. Is Dual-Energy CT: Indicates that this series uses dual-energy CT technology, capturing images at two different energy levels to differentiate materials based on their attenuation properties.
13. Is 3D Reconstruction: Indicates that this series includes three-dimensional reconstructed images from CT data, providing detailed anatomical visualization useful for surgical planning or assessment of complex structures.
14. Is Localizer: Indicates that this series is a localizer scan, used as initial scans for planning subsequent imaging sequences by providing anatomical references and orientation.
15. Is Helical: Indicates that this series uses helical (or spiral) CT scanning technique, where the X-ray tube rotates continuously around the patient while the table moves through the gantry, resulting in faster scan times and improved image quality.
---
Now, proceed to analyze the provided DICOM headers, explain your reasoning for each positively matched classifier, and output the list accordingly. Remember to include only the classifiers that are positively identified based on the data from `SeriesDescription`, `StudyDescription`, and `ProtocolName`. Be thorough and cautious of confounding factors. Do not hallucinate. Do not mention unless you are sure. You are given the study description just for context, but only grade the Boolean classifiers on the given series, as there are multiple series per study. Thank you.
RETURN THE OUTPUT IN THE EXAMPLE FORMAT AS SHOWN, WITHOUT ANY ADDITIONAL COMMENTARY!!!!
<EXAMPLE OUTPUT>
Output:
- Has Contrast
- Is Abdomen CT
Explanation:
- Has Contrast because the SeriesDescription includes 'Contrast', indicating that contrast was used.
- Is Abdomen CT because the SeriesDescription contains 'Abdomen', indicating that this is an abdominal CT scan.
</EXAMPLE OUTPUT>
<END OF PROMPT>
<INPUT>
<DICOM_HEADERS_HERE>
</INPUT>
<OUTPUT>
Please see video attachment for examples of outputs and explanations (11:07 to 13:02).
PHI and Cybersecurity Attack Removal Prompt 🔗
Below is the prompt used by the LLM to remove PHI and cybersecurity threats from the source data before sending to the cloud for processing and/or logging
You will be provided with text snippets that are Series Descriptions, Protocol Names, or Study Descriptions from medical records. These snippets may contain Personal Health Information (PHI), Personally Identifiable Information (PII), and cybersecurity attack code. Your task is to sanitize each text snippet by removing any PHI, PII, and malicious code.
Instructions:
Remove any PHI/PII, including but not limited to:
Names of individuals (e.g., patients, doctors, operators)
Dates directly associated with a person (e.g., birth dates, exam dates)
Unique identifiers (e.g., Medical Record Numbers, Patient IDs, Social Security Numbers)
Contact information (e.g., phone numbers, email addresses, physical addresses)
Remove any cybersecurity attack content, such as:
SQL injection code
Cross-site scripting (XSS) scripts
Malicious code snippets
Format string specifiers used maliciously (e.g., %s, %x, %n)
Do not remove any general medical terms, procedural details, or non-identifying information relevant to the description.
Ensure the final text is clear, coherent, and maintains proper grammar.
Only output the sanitized text. Do not include any additional commentary, explanations, or notes.
Examples:
Input:
Series Description: PRONE SCOUT - Patient Name: John A. Smith; DOB: 07/14/1965; ID: 123456
Output:
Series Description: PRONE SCOUT
Input:
Study Description: CT, COLONOGRAPHY SCREE <script>alert('You have been hacked');</script>
Output:
Study Description: CT, COLONOGRAPHY SCREE
Input:
Protocol Name: 4.6 COLONOSCOPY (ACRIN) %s%s%s%s%s
Output:
Protocol Name: 4.6 COLONOSCOPY (ACRIN)
Input:
Series Description: AXIAL T1 POST-CONTRAST - Operator: Dr. Emily Watson; MRN: 789012; Contact: (555) 987-6543
Output:
Series Description: AXIAL T1 POST-CONTRAST
Input:
Protocol Name: BRAIN MRI ROUTINE - SELECT * FROM Users WHERE '1'='1';
Output:
Protocol Name: BRAIN MRI ROUTINE
Now, please process the following text accordingly:
[Insert Unsafe String Here]
Note: Remember to only output the sanitized text with no additional commentary.
Question 1: Does FDA agree our anti-hallucination prompting and explainability strategies are sufficient?
Test Dataset Inventory 🔗
The below describes how the test set is created. The test set will be used for standalone performance testing of the device.
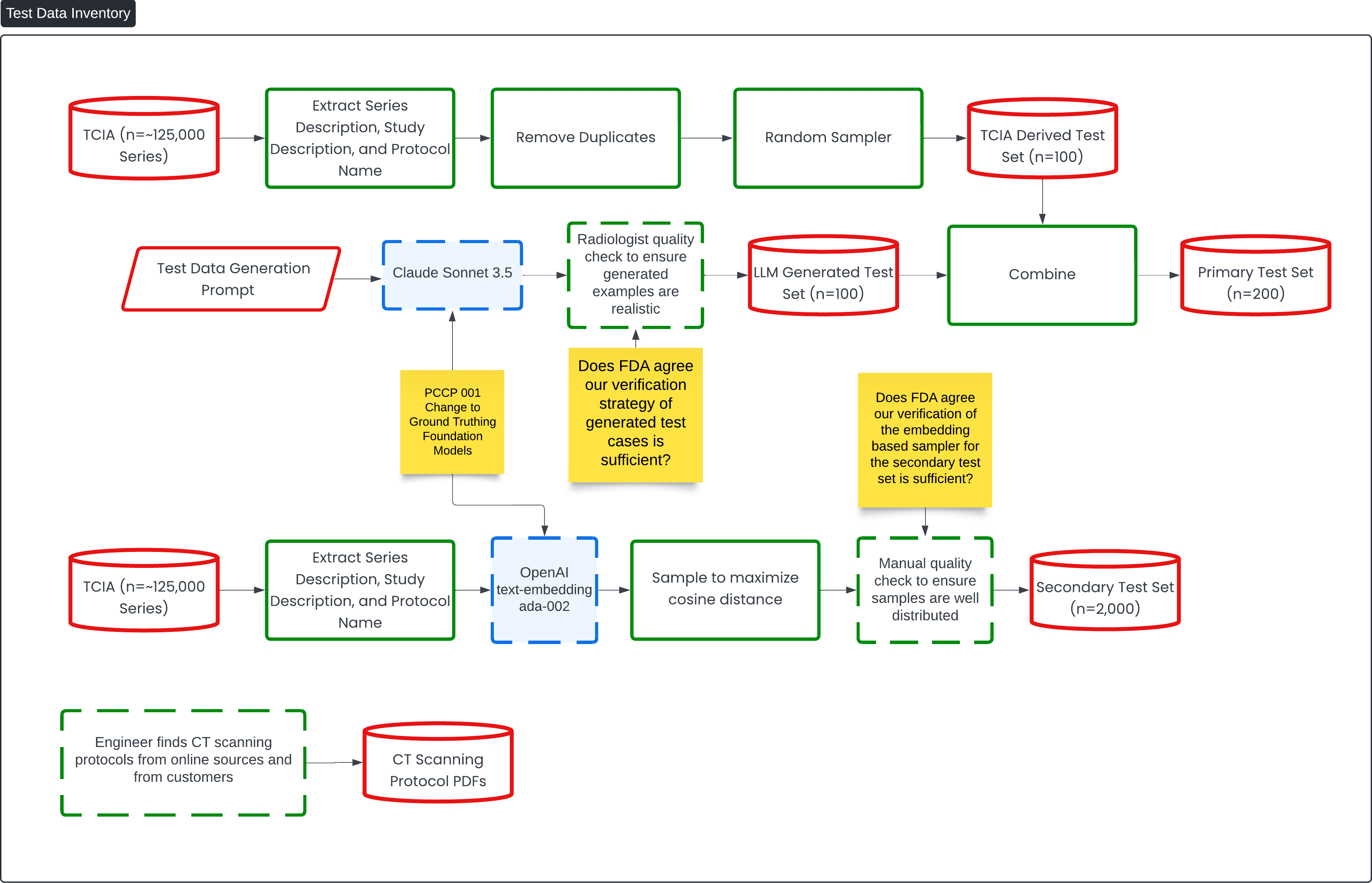
This diagram titled "Test Data Inventory" outlines the process of creating and verifying test sets for the Contrast Llama device, leveraging large data sets and machine learning models for classification tasks.
The flow begins with a large dataset of ~125,000 series from the TCIA (The Cancer Imaging Archive), from which the series description, study description, and protocol name are extracted. Duplicates are removed, and a random sample is taken to create a TCIA-derived test set of 100 samples. Additionally, an LL-generated test set (n=100) is created using a Test Data Generation Prompt run through the Claude Sonnet 3.5 model. A radiologist quality check is performed to ensure the generated examples are realistic.
These two datasets (the TCIA-derived set and the LLM-generated set) are combined to form the Primary Test Set (n=200). The goal is to check whether the FDA agrees that the verification strategy used for generating these test cases is sufficient.
A secondary process creates a Secondary Test Set (n=2,000) using the same TCIA data, which is again extracted and then processed using OpenAI's text-embedding model (ada-002). The secondary set is sampled to maximize cosine distance between examples, ensuring diverse samples. A manual quality check is performed to ensure the samples are well distributed.
The diagram also shows how CT scanning protocols are incorporated, with engineers sourcing protocols from online sources and customers to add variability and realism to the test sets. This enhances the overall testing strategy for the Contrast Llama device. Key review points, indicated in yellow, focus on obtaining FDA feedback on the adequacy of the verification strategies for both the generated test cases and the embedding-based sampler.
Question 2: Does FDA agree our verification strategy for generated test cases and of the embedding sampler for the secondary test set is sufficient?
Test Generation Prompt 🔗
The following is the prompt used to generate part of the test set.
Generate a comprehensive list of example CT scan protocols that includes the following for each entry:
Study Description: A concise title summarizing the entire CT examination.
Series Descriptions: Specific titles for each image series acquired during the scan.
Protocol Name: The technical name or designation of the scanning protocol used by radiology departments.
Your list should cover all types of common and uncommon CT protocols across various anatomical regions and clinical indications. Include variations based on:
Anatomical Regions: Head, neck, chest, abdomen, pelvis, spine, extremities.
Specialized Studies: Angiography (CTA), perfusion studies, virtual colonoscopy, cardiac CT, dental CT, low-dose screening CT, trauma imaging.
Contrast Usage: Both contrast-enhanced and non-contrast studies.
Contrast Phases: Arterial phase, venous phase, delayed phase, equilibrium phase.
Patient Positions and Techniques: Prone, supine, decubitus positions; inspiratory and expiratory scans; high-resolution techniques.
Age-Specific Protocols: Pediatric and adult protocols.
Functional and Dynamic Studies: CT perfusion, 4D CT scans, dynamic airway studies.
Format the output as a numbered list, and for each protocol, present the information in this structure:
Study Description: [Study title]
Series Descriptions:
[Series 1 title]
[Series 2 title]
[Series 3 title] (add as many as applicable)
Protocol Name: [Protocol designation]
Ensure that your examples reflect a variety of clinical scenarios and imaging needs. Provide at least 30 different protocols to encompass the diversity of CT imaging.
Foundation Model Training Description 🔗
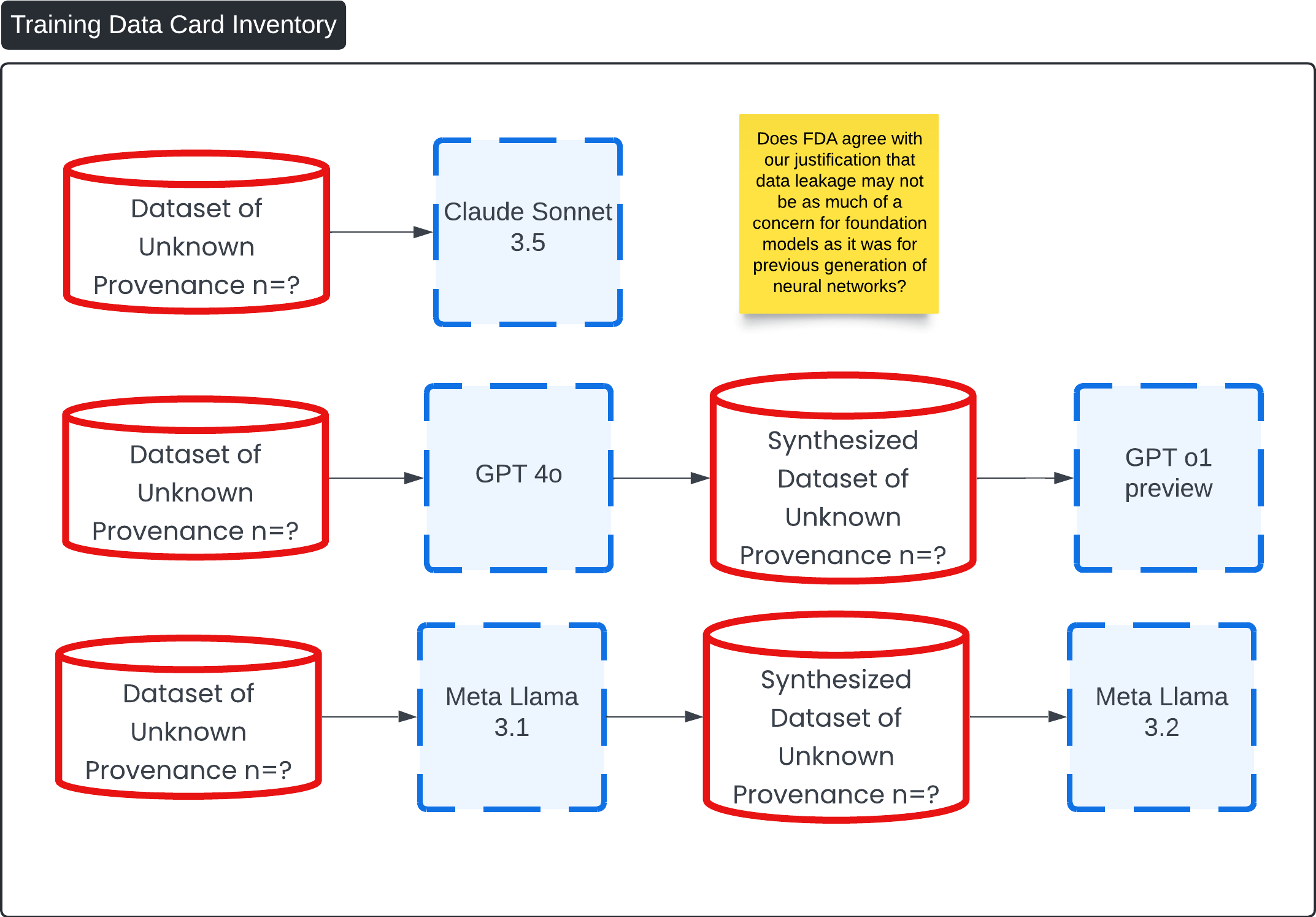
This diagram, titled "Training Data Card Inventory," outlines the process of synthesizing datasets using various foundation models, all of which are based on datasets of unknown provenance (n=?).
Data leakage is a critical issue in machine learning that can lead to overly optimistic performance estimates and poor generalization to new data. It typically occurs when information not available during deployment inadvertently influences the training process. However, in the context of foundation models like large language models (LLMs) used in the 'Contrast Lama' device, the risk of data leakage is significantly mitigated compared to previous generations of neural networks.
1. Nature of Foundation Models
- Extensive Training Data: Foundation models are trained on vast and diverse datasets that encompass a wide range of language usage across different domains. This extensive training reduces the likelihood that any specific data point, such as those used in validation or testing, would unduly influence the model's behavior.
- Generalized Learning: LLMs capture general language patterns rather than memorizing specific data instances. This means they are less prone to overfitting on particular datasets, a common consequence of data leakage.
2. Controlled Input and Output Mechanisms
- Structured Prompts and Outputs: The device employs carefully crafted prompts that instruct the LLM to provide boolean classifications along with explanations of its reasoning. This controlled interaction reduces the possibility of the model accessing or revealing unintended information.
3. Robust Verification and Validation Strategies
- Multi-Level Testing: The device's performance is validated using both human annotators and separate LLMs not involved in the initial training or operation. This multi-faceted approach ensures that any anomalies due to data leakage are detected.
- Ongoing Surveillance and Data Drift Mitigation: Nightly tests and continuous monitoring are implemented to detect and address any deviations in the model's behavior promptly. This proactive strategy helps maintain the integrity of the model over time.
Conclusion
Given the generalized nature of foundation models, their training on extensive and diverse datasets, and the specific risk control measures implemented in the 'Contrast Lama' device, data leakage is less of a concern compared to previous generations of neural networks. The device's design ensures that it operates within a controlled environment, with rigorous verification processes that uphold data integrity and model reliability. Therefore, we believe that the FDA can agree with the justification that data leakage risks are significantly mitigated in this context.
Question 3: Does FDA agree with our justification that data leakage may not be as much of a concern for foundation models as it was for previous generation of neural networks?
Manual Ground Truthing Description 🔗
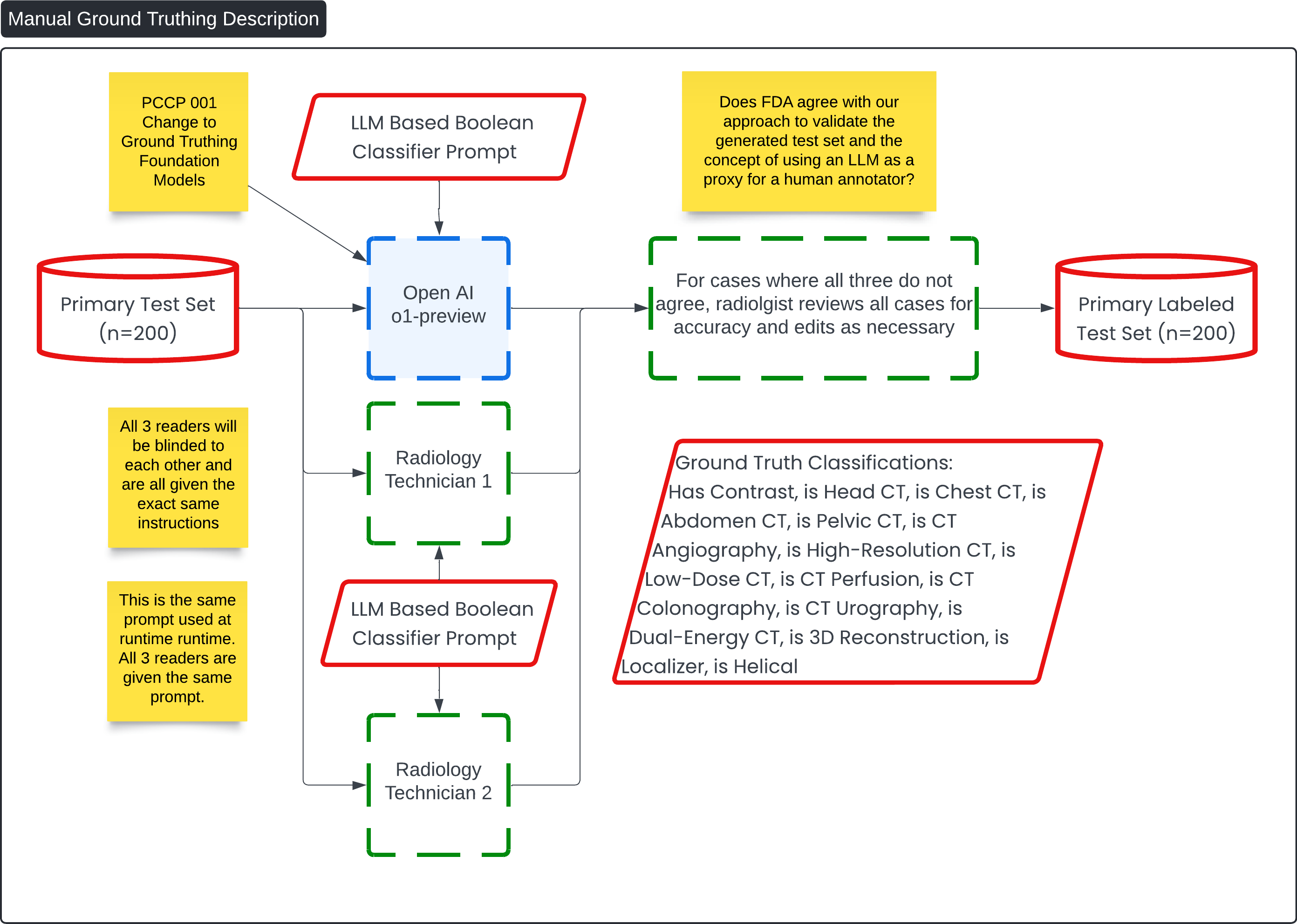
This diagram, titled "Manual Ground Truthing Description," outlines the process for labeling the Primary Test Set (n=200) using a combination of radiology technician inputs and an AI-driven approach. The goal is to create a labeled test set with high accuracy, leveraging both human and machine insights.
Key Process Flow: 🔗
-
Primary Test Set Input (n=200):
- This is the initial dataset that will undergo classification and ground truthing. It includes various imaging cases such as CT scans that need to be labeled with specific attributes.
-
LLM-Based Boolean Classifier Prompt:
- The same prompt used during runtime is also employed here for consistency. It is fed into both an LLM (OpenAI o1-preview) and two radiology technicians, all of whom are blinded to each other's results to ensure unbiased results.
-
OpenAI o1-preview:
- This LLM is tasked with classifying the test set using the provided Boolean classifier prompt. It outputs classifications such as whether the scan has contrast, or if it is a head CT, chest CT, etc.
-
Radiology Technicians (Reader 1 and Reader 2):
- Two radiology technicians also receive the same prompt and are tasked with manually classifying the cases. They will review the cases independently, and their results will be compared with the LLM's output.
-
Validation of Results:
- If all three readers (the LLM and two technicians) agree on the classification, the result is accepted. If there is any discrepancy, a radiologist reviews all cases to ensure accuracy, making necessary edits and corrections.
-
Final Ground Truth Classifications:
- Once all discrepancies are resolved, the final ground truth classifications are recorded, including key tags like "Has Contrast," "Is Head CT," "Is Abdomen CT," and others. These classifications are crucial for the downstream medical imaging workflows.
-
Primary Labeled Test Set (n=200):
- The final output is a labeled test set that has been validated through this hybrid approach, combining LLM output with human expertise. This test set can now be used for further model training, verification, or clinical usage.
Example Ground Truth 🔗
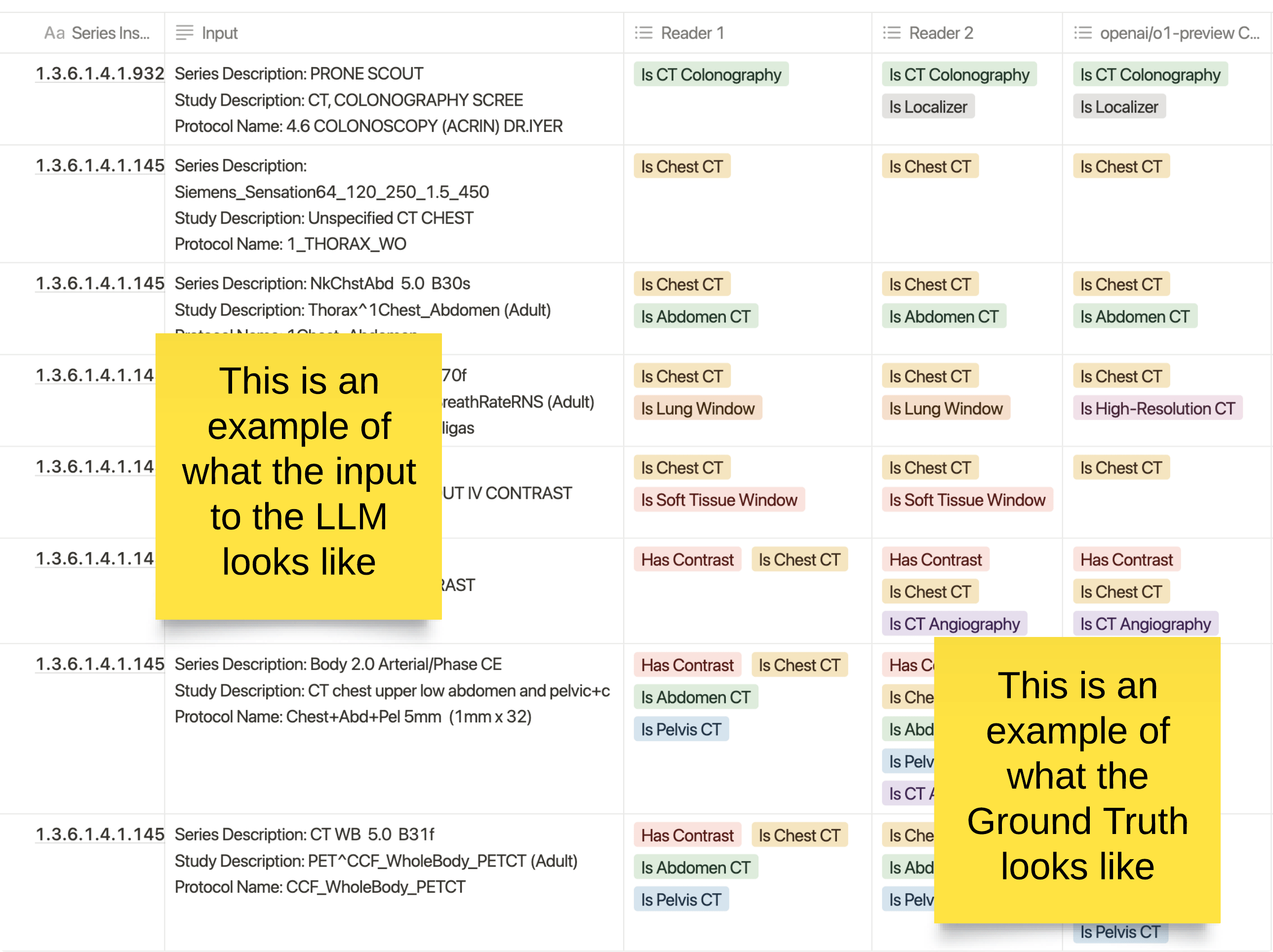
This image provides a visual example of the process used in creating ground truth for medical imaging classifications, specifically through the use of human annotators and a large language model (LLM).
Key Elements: 🔗
- Input to the LLM: The left column represents what is fed into the LLM for classification. It includes various metadata related to CT scans, such as the series description, study description, and protocol name. This information provides context to the LLM for making predictions about the type of scan.
- Human Readers: Columns for Reader 1 and Reader 2 represent manual classifications provided by radiology technicians. Each technician is blinded to the other’s results, providing an independent assessment of the scan. They classify whether the scan is a "Chest CT," "Abdomen CT," "Has Contrast," "CT Colonography," or other relevant attributes.
- LLM Predictions (OpenAI o1-preview): The final column shows the classifications predicted by the LLM (OpenAI o1-preview) for the same input. These classifications are compared to the radiologists' annotations to ensure alignment or identify discrepancies.
The Ground Truth: 🔗
The ground truth is established when the results from the LLM and the two human readers are compared. In cases where all three (Reader 1, Reader 2, and the LLM) agree, the result becomes part of the ground truth. In cases of disagreement, a radiologist reviews the output to resolve any discrepancies, ensuring high-quality data for training and testing.
This process is designed to combine human expertise with AI to create a reliable labeled dataset for use in clinical applications, improving both the accuracy and scalability of medical imaging classification.
Question 4: What are FDA’s thoughts on the concept of using an LLM as a proxy for a human annotator?
Automatic Ground Truthing Description 🔗
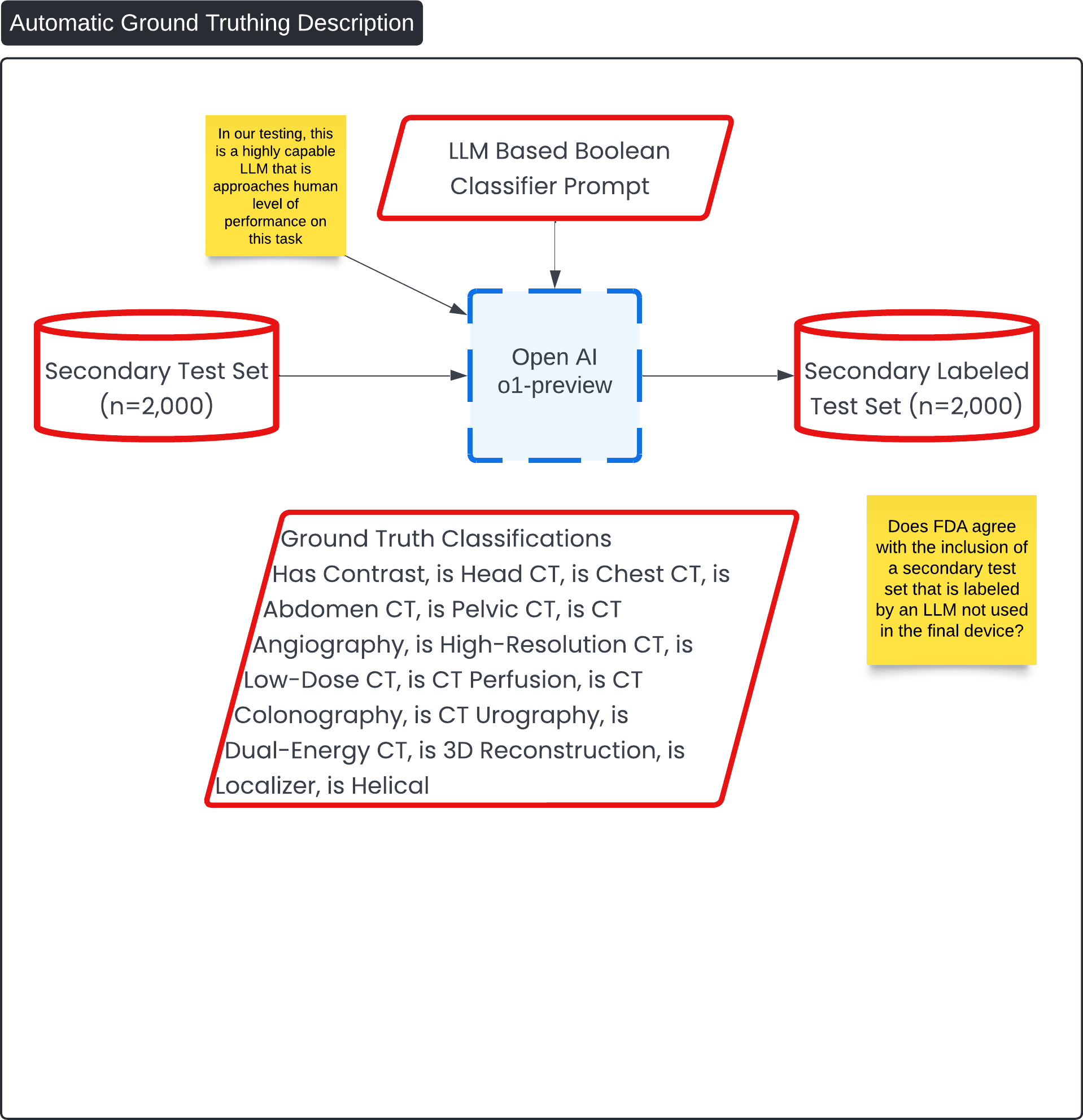
This diagram, titled "Automatic Ground Truthing Description," presents the process for generating a Secondary Labeled Test Set (n=2,000) using an AI-driven approach. The goal is to create a labeled dataset with minimal human intervention, leveraging an LLM (Large Language Model) to automatically classify medical imaging data.
Key Process Flow: 🔗
-
Secondary Test Set (n=2,000):
- This dataset forms the initial input and consists of imaging data that requires classification into various categories.
-
LLM-Based Boolean Classifier Prompt:
- A Boolean classifier prompt is applied to the test set. This is the same prompt used during the manual ground truthing process, ensuring consistency in the classifications.
-
OpenAI o1-preview:
- The OpenAI o1-preview model is used to process the test set. This model is tasked with classifying each entry in the test set based on the provided Boolean classifier prompt. The model outputs classifications for various CT scan features (e.g., "Has Contrast," "Is Chest CT," "Is Abdomen CT").
-
Secondary Labeled Test Set (n=2,000):
- The output of the OpenAI model forms the Secondary Labeled Test Set, which contains the ground truth classifications for the data.
Question 5: Does FDA agree with the inclusion of a secondary test set that is labeled by an LLM not used in the final device?
Traditional ML Verification 🔗
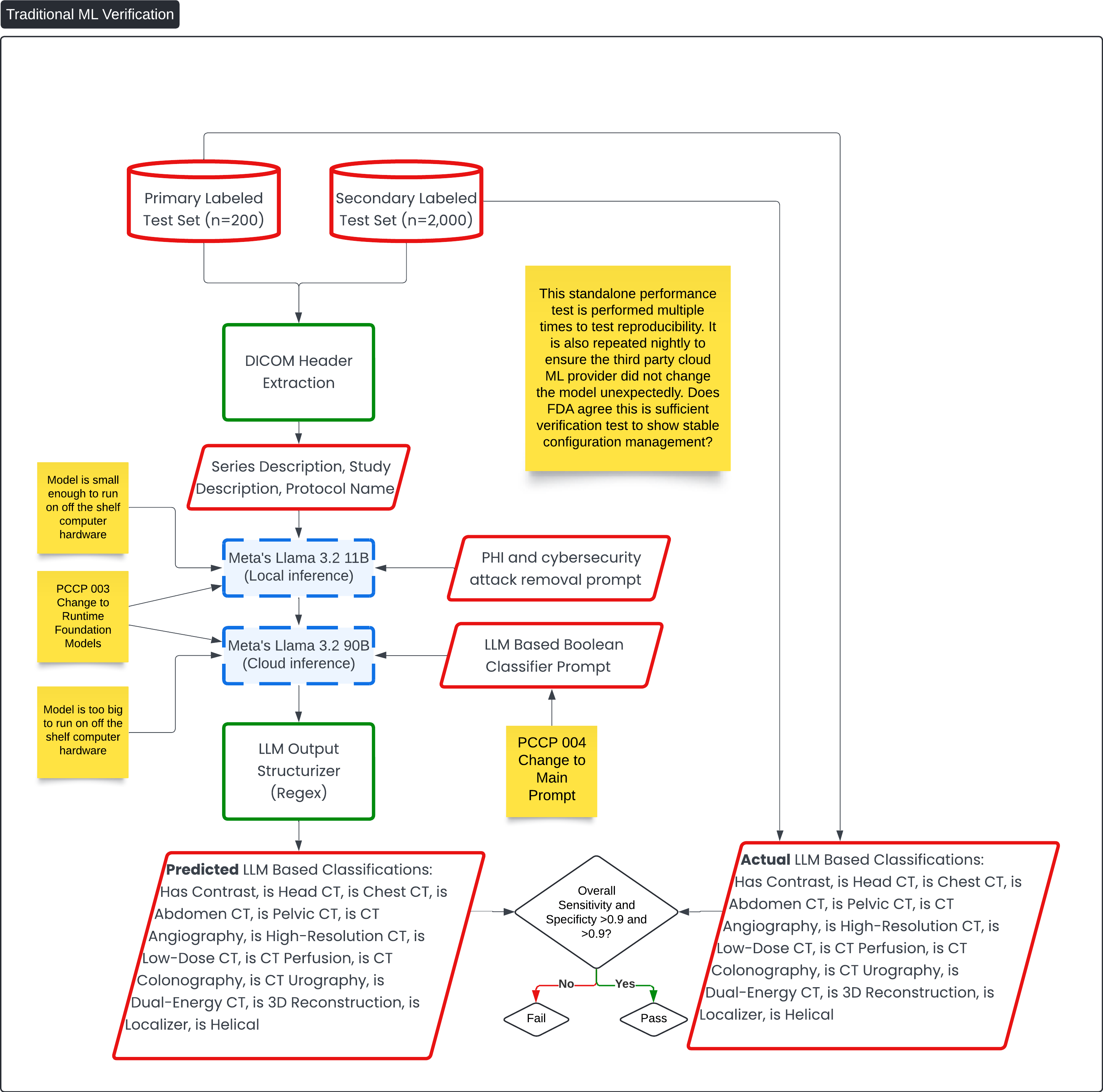
This diagram, titled Traditional ML Verification, details the verification process for the machine learning (ML) components of the Contrast Llama Device for accuracy compared to the ground truth and reproducibility compared to subsequent runs to itself.
Key Process Flow: 🔗
-
Primary Labeled Test Set (n=200) and Secondary Labeled Test Set (n=2,000):
- These labeled test sets are used to verify the accuracy of the ML models. The test sets contain ground truth data with attributes such as "Has Contrast," "Is Head CT," and "Is Pelvic CT."
-
DICOM Header Extraction:
- This component extracts key metadata, such as series description and study description, from the DICOM files. This data serves as input for the ML models.
-
ML Models - Meta's Llama 3.2:
- Local Inference: The smaller model (Meta's Llama 3.2 11B) runs locally on available hardware. This runs the PHI and cybersecurity removal prompt.
- Cloud Inference: The larger model (Meta's Llama 3.2 90B) runs on cloud infrastructure because it is too large to run on standard hardware. This runs the main boolean classification prompt for the main function of the device.
-
LLM Output Structurizer (Regex):
- After the models generate predictions, the output is structured into a machine-readable format using regular expressions.
-
Predicted vs. Actual LLM-Based Classifications:
- The predicted classifications generated by the model (e.g., "Has Contrast," "Is Head CT," "Is Pelvic CT") are compared to the actual, labeled classifications from the test sets.
- The overall sensitivity and specificity are calculated. The goal is to achieve both values greater than 0.9. If the test passes, the model is validated; if not, adjustments are needed.
-
Ongoing Verification for Stability:
- Nightly Testing: The test is repeated nightly to verify that the third-party cloud ML provider has not inadvertently changed the model. This ensures that the model remains stable over time.
- The process incorporates multiple performance tests to validate reproducibility and model integrity. This verifies risk control measures, such as setting the temperature to 0, which reduces the likelihood of non-deterministic outputs and hallucinations.
Question 6: This standalone performance test is performed multiple times to test reproducibility. It is also repeated nightly to ensure the third party cloud ML provider did not change the model unexpectedly. Does FDA agree this is sufficient verification test to show stable configuration management?
ML Verification View for PHI and Security Threat Scrubbing 🔗
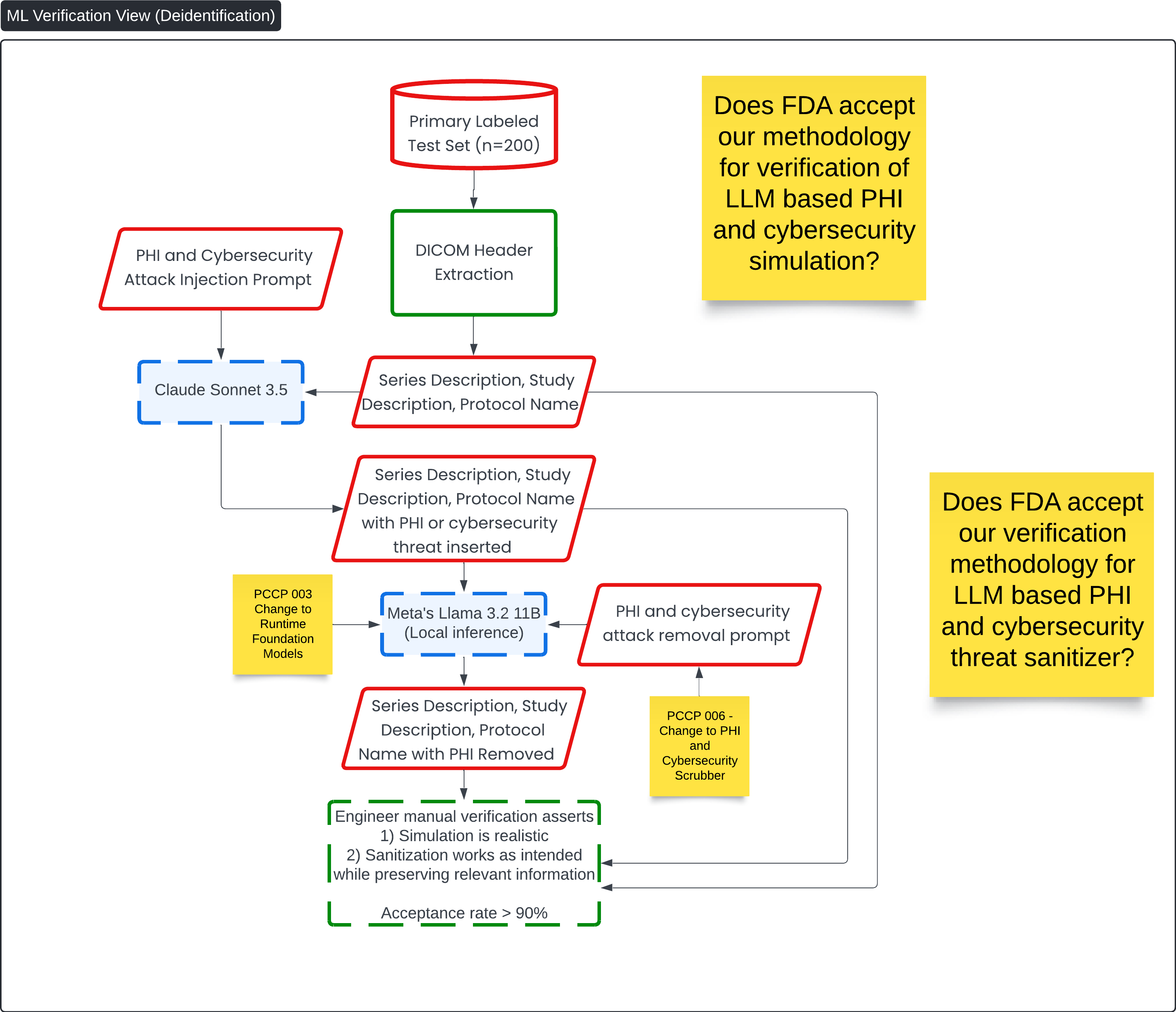
This diagram, titled ML Verification View (Deidentification), focuses on the verification process for PHI (Protected Health Information) and cybersecurity attack removal in the Contrast Llama system. It outlines the methodology for simulating PHI and cybersecurity threats, then verifying that the LLM (Meta's Llama 3.2 11B) effectively sanitizes this sensitive information while preserving relevant details needed for medical image analysis.
Key Process Flow: 🔗
-
Primary Labeled Test Set (n=200):
- The test set is prepared with labeled medical images and metadata, serving as the basis for the PHI and cybersecurity threat simulation.
-
PHI and Cybersecurity Attack Injection Prompt:
- The Claude Sonnet 3.5 model injects simulated PHI and cybersecurity threats into the DICOM metadata (series description, study description, and protocol name). This step helps simulate real-world scenarios where sensitive data or malicious code could be present.
-
DICOM Header Extraction:
- The DICOM header information is extracted, and the relevant metadata is modified with injected threats or PHI.
-
Meta's Llama 3.2 (Local Inference):
- The modified metadata, now containing PHI or cybersecurity threats, is processed by Meta's Llama 3.2 11B, which uses a PHI and cybersecurity attack removal prompt to sanitize the data.
-
PHI and Cybersecurity Removal:
- After processing, the model outputs metadata where the PHI or cybersecurity threats have been removed. The sanitized output retains important medical information necessary for clinical workflows.
-
Engineer Verification:
- A manual verification process is carried out by an engineer who ensures two things:
- The simulation is realistic: The injected PHI and cybersecurity threats are representative of real-world data.
- The sanitization is effective: The LLM successfully removes the PHI and threats while preserving the integrity of relevant information needed for analysis.
The acceptance rate for this process must be greater than 90% to confirm successful sanitization.
Question 7: Does FDA accept our methodology for verification of LLM-based PHI, cybersecurity simulation and cybersecurity threat sanitizer?
- A manual verification process is carried out by an engineer who ensures two things:
Non ML Verification View 🔗
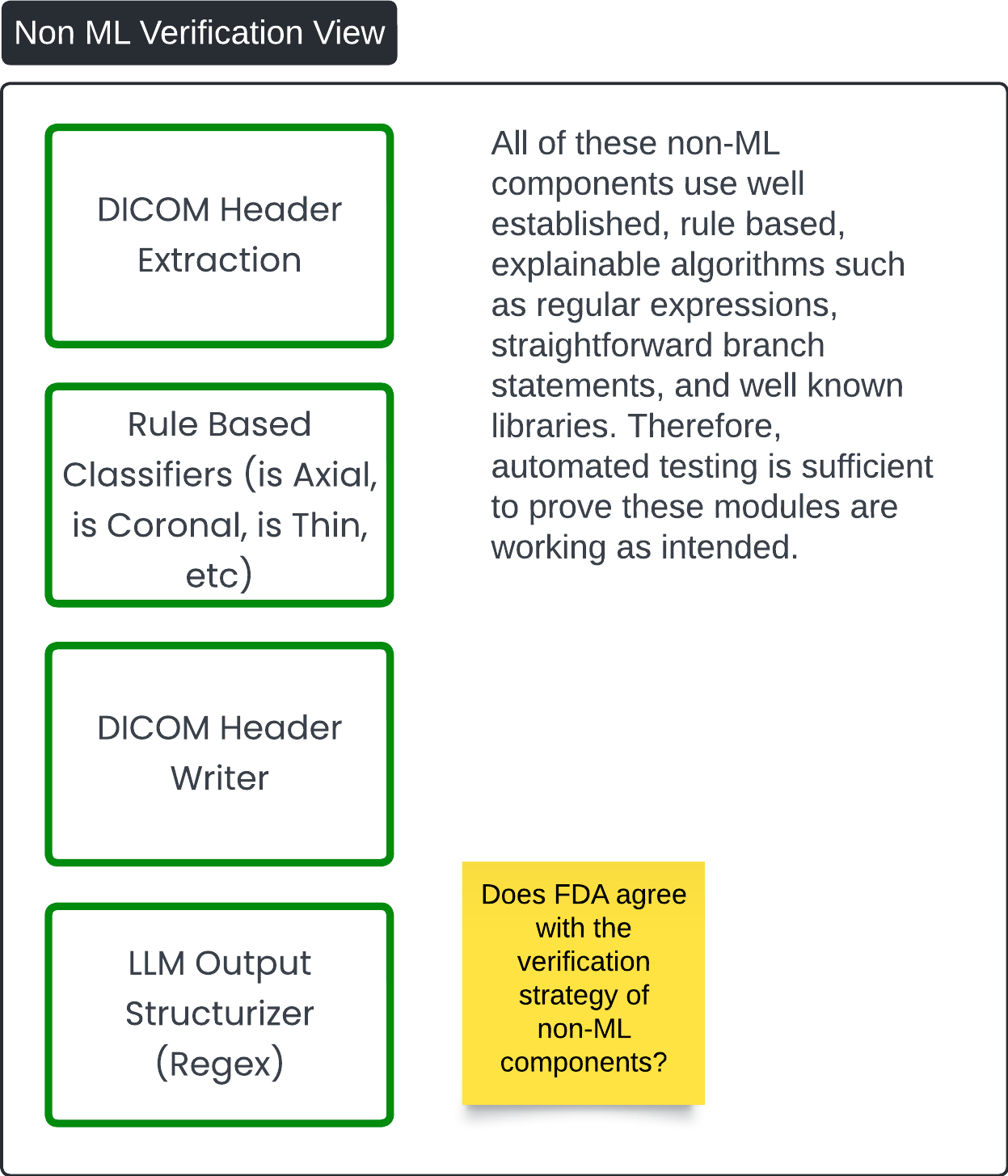
This diagram, titled Non-ML Verification View, outlines the verification strategy for non-machine learning (non-ML) components within the Contrast Llama system. The diagram highlights key components that rely on rule-based and explainable algorithms, and presents the justification for using automated testing to verify their functionality.
Key Components: 🔗
-
DICOM Header Extraction:
- This component extracts metadata from DICOM files, focusing on the DICOM headers. It uses straightforward, rule-based logic to retrieve and organize necessary information.
-
Rule-Based Classifiers:
- These classifiers, which determine attributes like whether the scan is axial, coronal, or thin, are based on established, explainable algorithms (e.g., regular expressions or decision trees). These algorithms are well-known and predictable, making them easier to validate through automated tests.
-
DICOM Header Writer:
- The DICOM Header Writer adds new information (tags) to the original DICOM files. It follows predefined rules to ensure the original pixel data remains unchanged while new tags are added accurately.
-
LLM Output Structurizer (Regex):
- This component uses regular expressions to structure the output of the LLM in a machine-readable format, such as JSON or similar. It ensures that the output is cleanly organized and follows consistent formatting rules.
Verification Strategy: 🔗
-
Automated Testing:
- The non-ML components rely on deterministic algorithms that are well-established and explainable. As a result, automated tests can sufficiently verify that these modules are functioning as intended. Given that these components are not reliant on data-driven, probabilistic models, their behavior is predictable and can be thoroughly tested using established test cases and libraries.
This diagram reinforces that non-ML components, due to their explainability and reliance on established algorithms, can be reliably tested using automation, ensuring their robustness and reliability within the overall system.
Automated ML Verification 🔗
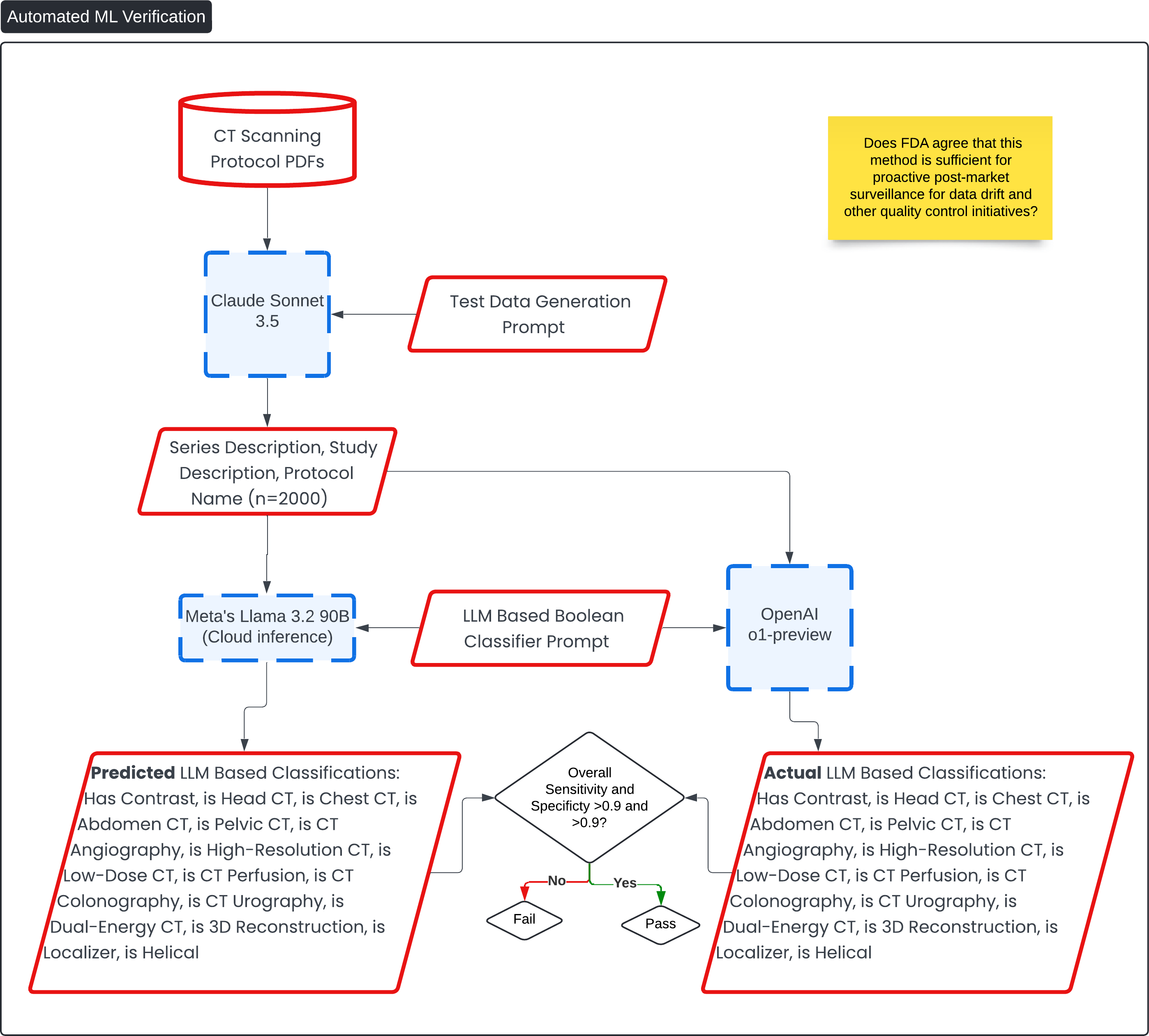
This diagram, titled Automated ML Verification, outlines the process for automatically verifying the performance of the Contrast Llama system using large language models (LLMs) and generated test data. The goal of this verification process is to ensure that the model maintains high accuracy and robustness for ongoing quality control and post-market surveillance.
Key Process Flow: 🔗
-
CT Scanning Protocol PDFs:
- A dataset of CT scanning protocols is used as the foundational input for generating test data. These protocols define the series description, study description, and protocol name relevant to medical imaging.
-
Test Data Generation Prompt:
- Using Claude Sonnet 3.5, a test data generation prompt is applied to the CT protocols. This creates a dataset of 2,000 entries with detailed metadata for testing.
-
LLM-Based Classification:
- The metadata generated from the CT protocols is processed by Meta's Llama 3.2 90B (a cloud-based inference model), which applies a Boolean classifier prompt to classify the images based on key characteristics such as whether the image has contrast or is a head, chest, or pelvic CT scan.
-
Comparison with OpenAI o1-preview:
- The same data is run through OpenAI o1-preview to generate a second set of classifications using the same prompt. This provides a comparative validation process for the LLM-based classifications.
-
Verification:
- The predicted classifications generated by Meta’s Llama 3.2 90B are compared with the actual classifications derived from OpenAI o1-preview and the known ground truth data.
- The test evaluates whether the sensitivity and specificity are greater than 0.9. If the model passes, it proceeds to further verification steps; if it fails, adjustments are required.
Verification of Automated ML Verification 🔗
This diagram, titled Automated ML Verification Verification, outlines the process for verifying the test methodology described in Automated ML Verification.
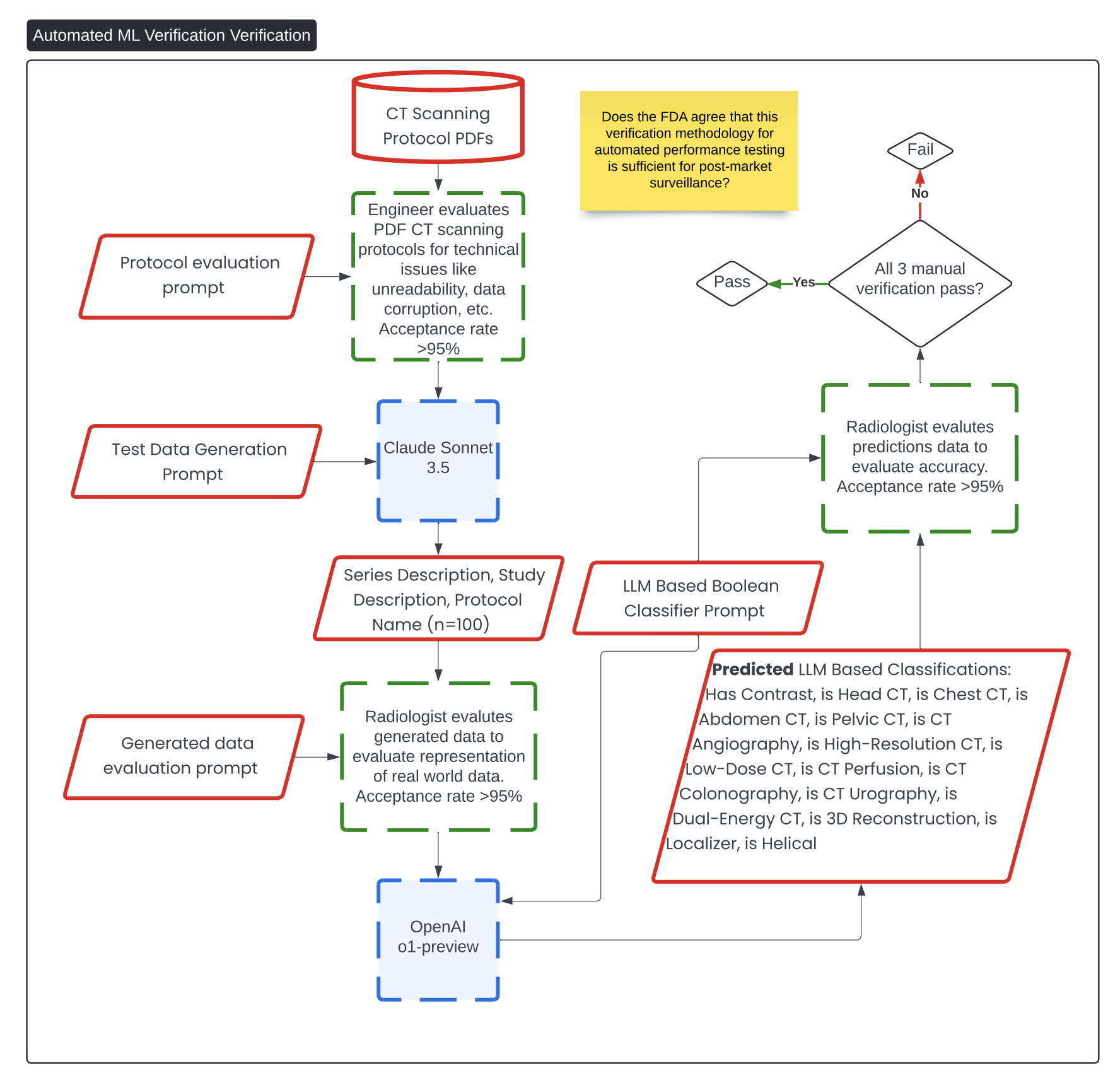
Question 8: Does FDA agree that the methodology described in the Automated ML Verification diagram is sufficient for proactive post-market surveillance for data drift and other quality control initiatives?
Predetermined Change Control Plan 🔗
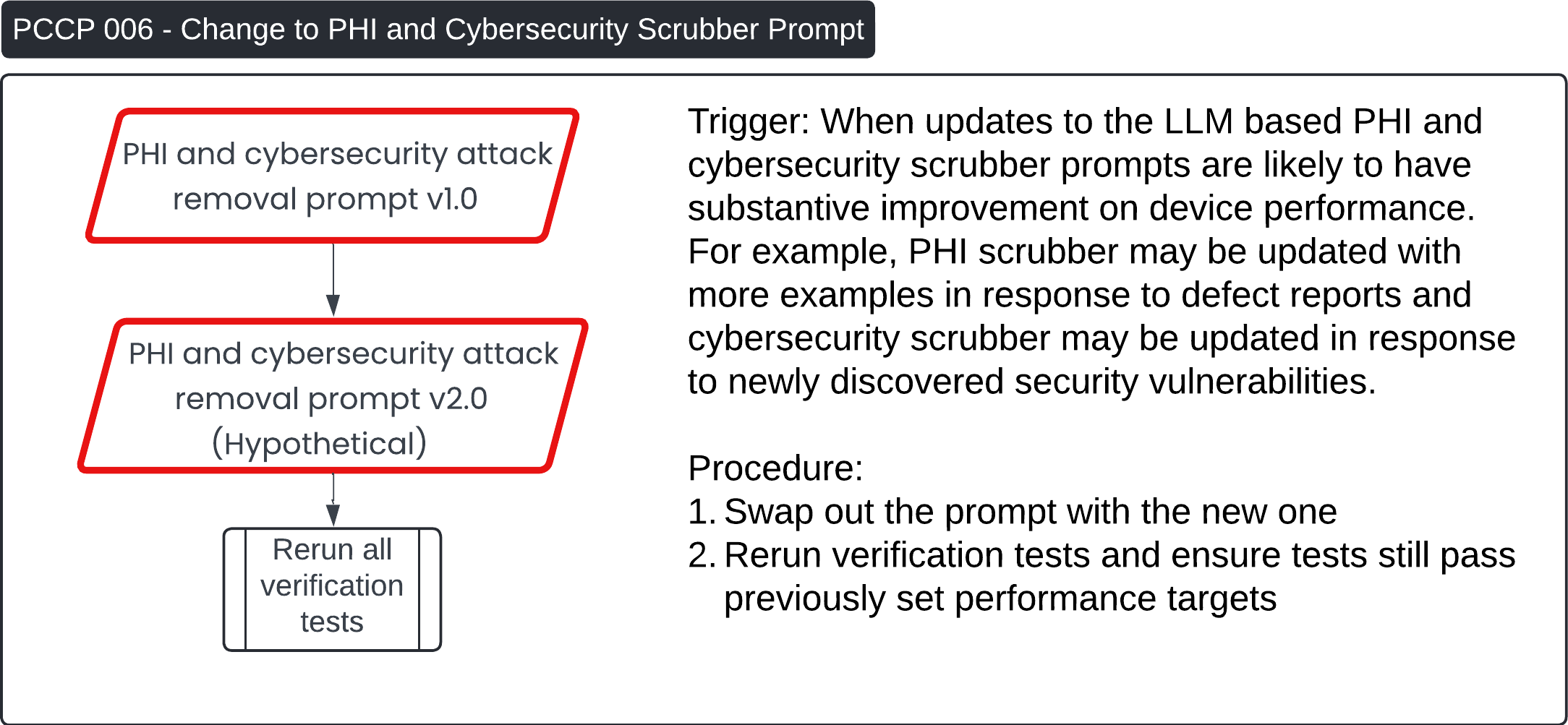
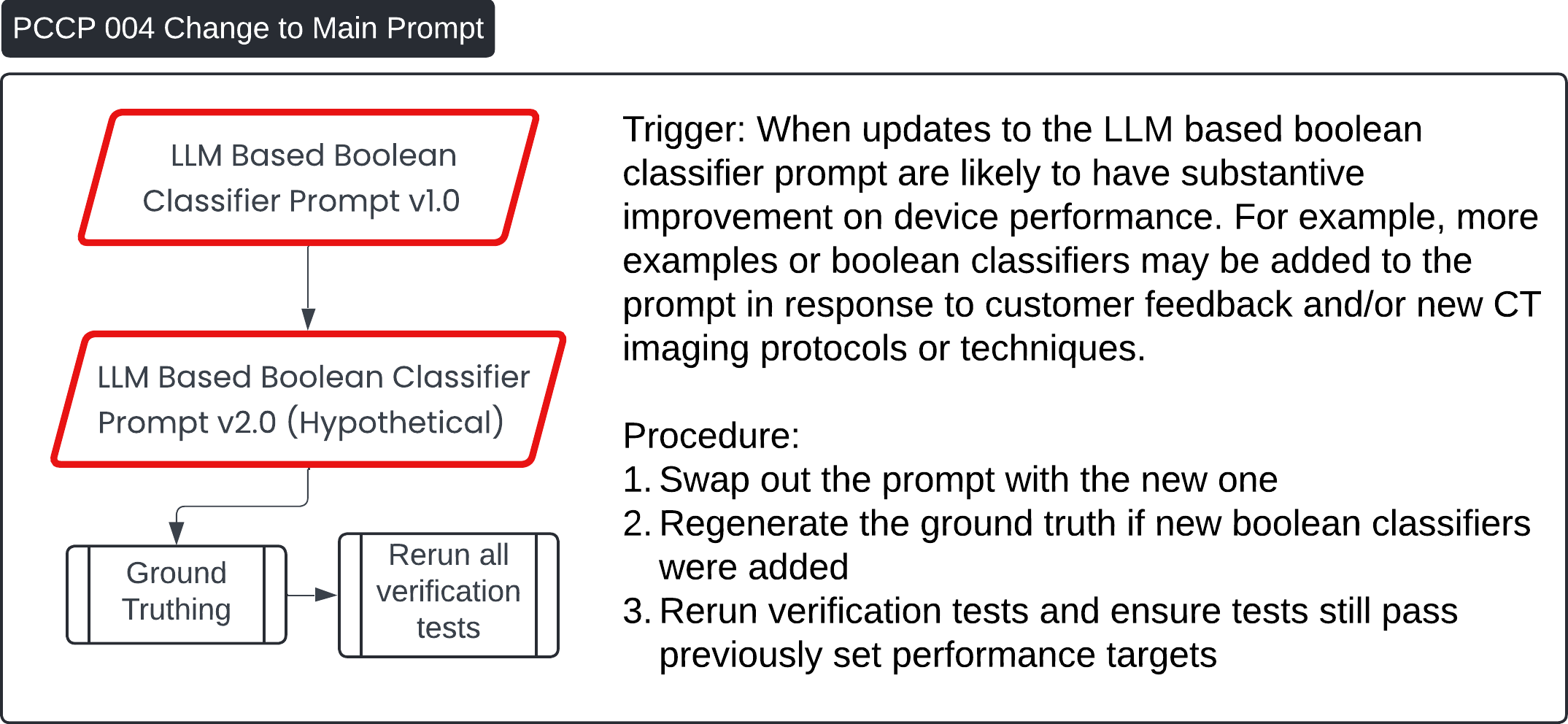
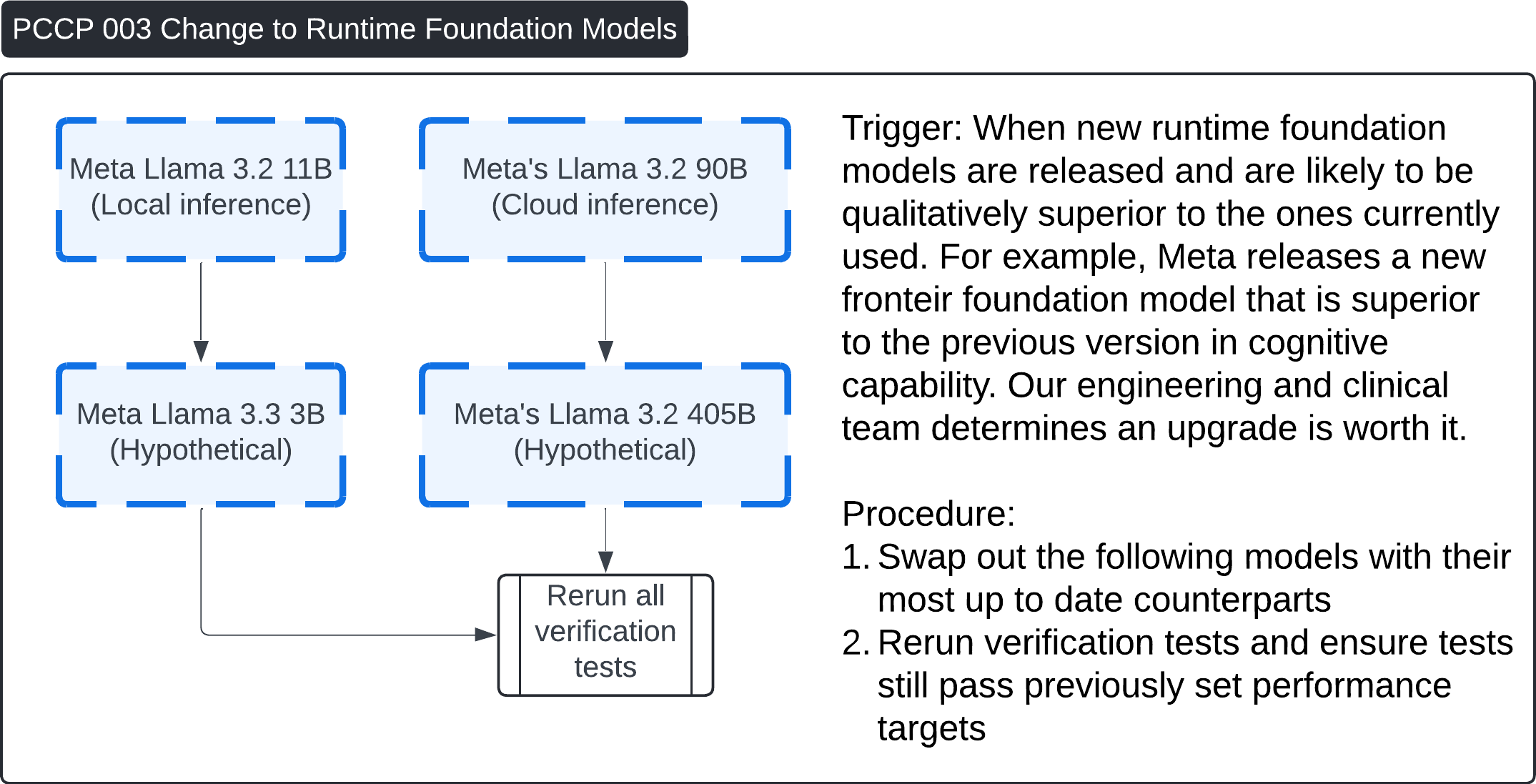
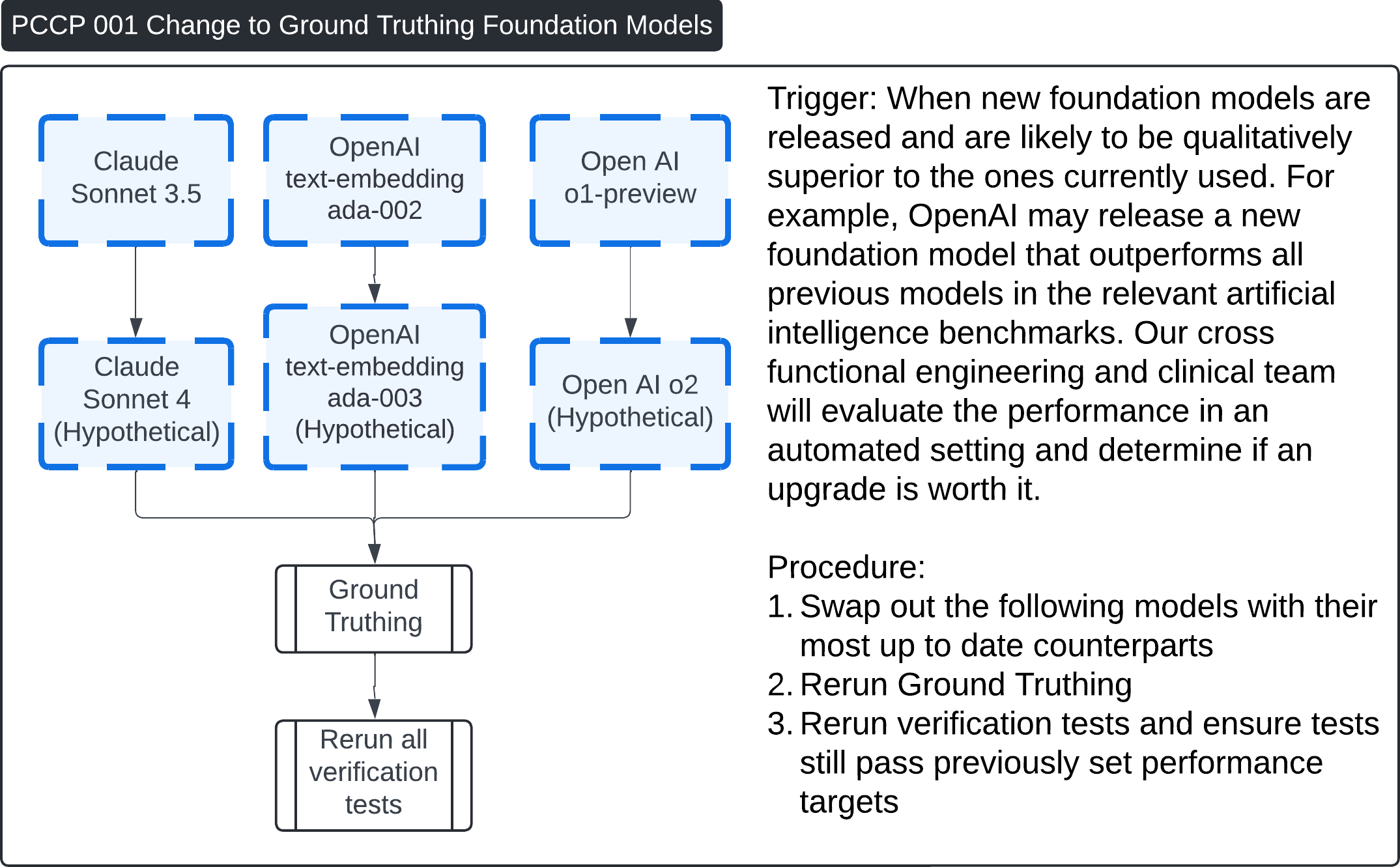
These four images describe various Post-Change Control Process (PCCP) workflows for updates to the Contrast Llama system:
-
PCCP 006 - Change to PHI and Cybersecurity Scrubber Prompt:
- This process is triggered when updates to the PHI (Protected Health Information) or cybersecurity scrubber prompt are made to enhance performance or address new vulnerabilities. The procedure involves swapping out the old prompt with the updated version (v2.0, hypothetical), followed by rerunning all verification tests to ensure performance remains within previously set targets.
-
PCCP 004 - Change to Main Boolean Classifier Prompt:
- This workflow outlines changes to the LLM-based Boolean classifier prompt (v1.0 to v2.0). The prompt may be updated based on customer feedback or new CT imaging protocols. After the update, the procedure includes regenerating the ground truth, rerunning all verification tests, and ensuring performance targets are still met.
-
PCCP 003 - Change to Runtime Foundation Models:
- This procedure is triggered when new, superior runtime foundation models are released (e.g., from Meta's Llama 3.2 11B to Llama 3.3 3B, hypothetical). The process involves upgrading to the most up-to-date model and rerunning all verification tests to ensure that the new model meets the original performance benchmarks.
-
PCCP 001 - Change to Ground Truthing Foundation Models:
- This describes the process when new ground truthing foundation models (e.g., Claude Sonnet or OpenAI models) are introduced. The workflow includes swapping out the foundation models, regenerating ground truth, and rerunning all verification tests to confirm performance remains consistent.
Each PCCP ensures that updates to the system—whether in prompts or foundation models—are carefully tested to maintain performance standards.
Question 9: Does FDA have any concerns about our PCCP?
Specific Questions 🔗
| Sponsor Question | Section | Topic |
|---|---|---|
| Question 1: Does FDA agree our anti-hallucination prompting and explainability strategies are sufficient? | Runtime Description | LLM Prompts |
| Question 2: Does FDA agree our verification strategy for generated test cases and of the embedding sampler for the secondary test set is sufficient? | Test Dataset Inventory | Testing and Training Datasets |
| Question 3: Does FDA agree with our justification that data leakage may not be as much of a concern for foundation models as it was for previous generation of neural networks? | Foundation Model Training Description | Testing and Training Datasets |
| Question 4: What are FDA’s thoughts on the concept of using an LLM as a proxy for a human annotator? | Manual Ground Truthing Description | Verification Testing Plan |
| Question 5: Does FDA agree with the inclusion of a secondary test set that is labeled by an LLM not used in the final device? | Automatic Ground Truthing Description | Verification Testing Plan |
| Question 6: This standalone performance test is performed multiple times to test reproducibility. It is also repeated nightly to ensure the third party cloud ML provider did not change the model unexpectedly. Does FDA agree this is sufficient verification test to show stable configuration management? | Traditional ML Verification | Verification Testing Plan |
| Question 7: Does FDA accept our methodology for verification of LLM-based PHI, cybersecurity simulation and cybersecurity threat sanitizer? | ML Verification View for PHI and Security Threat Scrubbing | Verification Testing Plan |
| Question 8: Does FDA agree that the methodology described in the Automated ML Verification diagram is sufficient for proactive post-market surveillance for data drift and other quality control initiatives? | Automated ML Verification | Verification Testing Plan |
| Question 9: Does FDA have any concerns about our PCCP? | Predetermined Change Control Plan | Predetermined Change Control Plan (PCCP) |