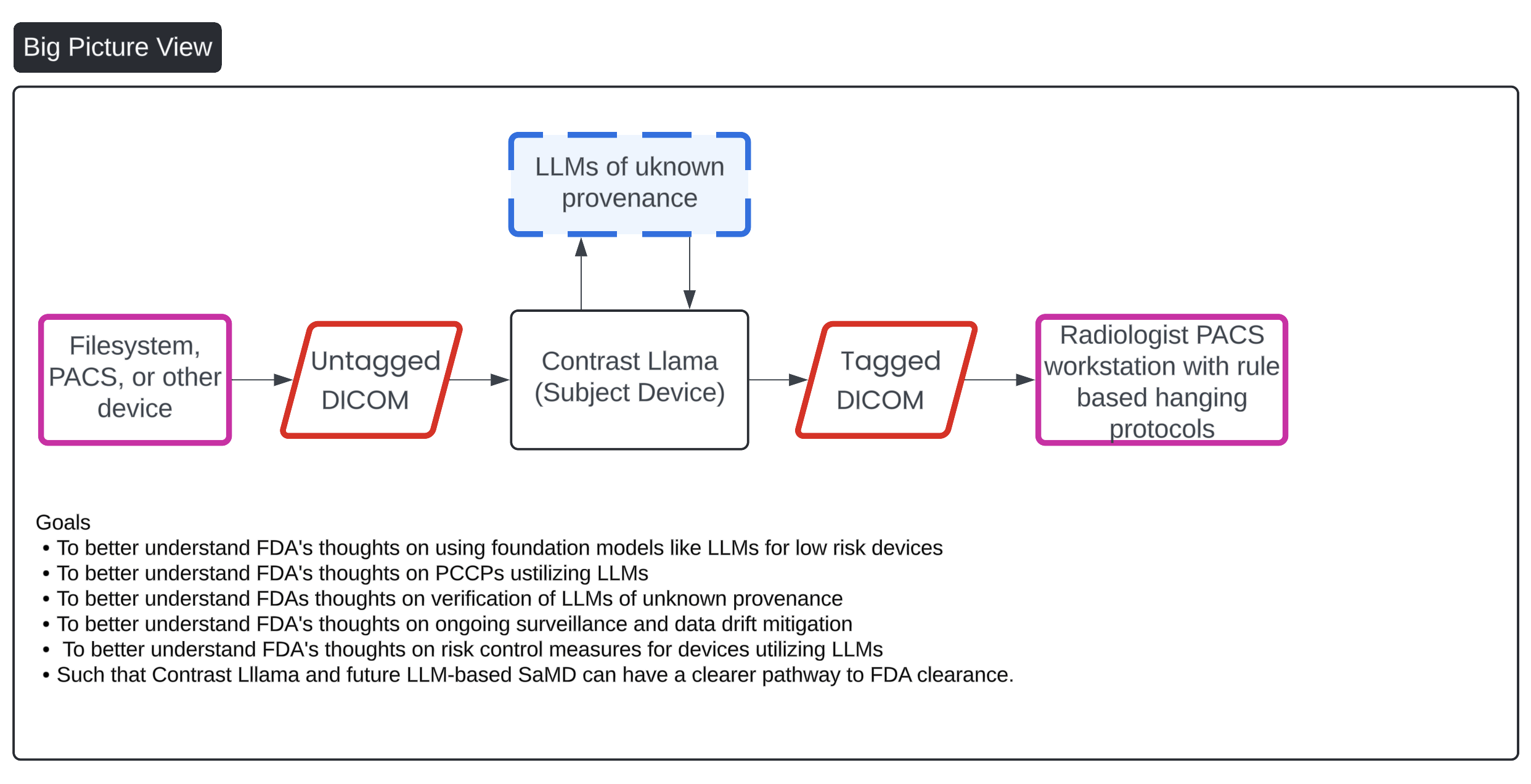
In recent years, the field of radiology has witnessed significant advancements driven by the development of foundation models. These models, which integrate vision and language capabilities, are designed to enhance the interpretation and analysis of medical images. The emergence of such models is transforming the landscape of medical imaging, offering new possibilities for clinical applications and research.
Radiology foundation models are built upon large-scale datasets and sophisticated architectures that enable them to handle complex multimodal information. A notable example is the RadFM, a pioneering model that leverages a vast dataset called MedMD, consisting of 16 million 2D and 3D medical scans. This dataset is unique for its inclusion of both 2D and 3D scans, providing a comprehensive resource for model training and evaluation.
The development of these models is supported by open-source platforms such as GitHub, where researchers and developers collaborate to refine and expand the capabilities of radiology foundation models. Repositories like Awesome Foundation Models in Medical Imaging and Awesome AI in Radiology curate a wealth of resources, including papers, datasets, and tools, facilitating the dissemination of knowledge and fostering innovation in the field.
One of the key challenges addressed by these models is the integration of vision-language architectures, as seen in projects like RadFound. This model is tailored for radiology and trained on an extensive dataset of over 8.1 million images and 250,000 image-text pairs, covering a wide range of organ systems and imaging modalities. RadFound introduces enhanced vision encoders and cross-modal learning designs, setting new benchmarks in radiology interpretation tasks.
The ongoing research and development efforts in radiology foundation models are not only advancing the technical capabilities of these systems but also promoting their practical applicability in clinical settings. By making codes, data, and model checkpoints publicly available, initiatives like RadFM and RadFound are paving the way for further exploration and application of AI in radiology.
As the field continues to evolve, the collaborative efforts on platforms like GitHub will play a crucial role in shaping the future of radiology foundation models, ultimately contributing to improved patient care and diagnostic accuracy.
Table of Contents 🔗
- Radiology Foundation Models on GitHub
- Development of RadFound
- RadFM: A Multimodal Approach
- Medical AI Research Foundations
- MONAI: A PyTorch-Based Framework
- Mayo-UGA RadOnc Foundation Models
- RadGenome-Chest CT Dataset
- Knowledge-Enhanced Vision-Language Pre-Training
- GPT-4V Evaluation
- PMC-LLaMA: An Open-Source Language Model
- Conclusion
- Key Datasets and Architectures in Radiology Foundation Models
- Large-Scale Medical Datasets
- Vision-Language Model Architectures
- Benchmarking and Evaluation Frameworks
- Open-Source Code Repositories
- Integration of AI in Clinical Practice
- Evaluation and Benchmarking of Radiology Foundation Models
- Evaluation Metrics and Methodologies
- Comparative Analysis of Foundation Models
- Benchmarking on Diverse Datasets
- Human and Automated Evaluation Techniques
- Challenges and Future Directions in Evaluation
Radiology Foundation Models on GitHub 🔗
| Model/Repository Name | Description | Key Features | Dataset(s) Used |
|---|---|---|---|
| RadFound | Vision-language model tailored for radiology. | Vision encoder capturing intra-image/local and inter-image/contextual information, cross-modal learning design. | 8.1 million images, 250,000 image-text pairs (RadVLBench). |
| RadFM | Multimodal radiology foundation model integrating 2D/3D scans. | Visually conditioned generative pre-training. | MedMD (16 million 2D/3D scans), RadMD (3 million image-text pairs). |
| Medical AI Research Foundations | Open-source models for chest X-ray and pathology. | Non-diagnostic models generating representations for medical images. | Public clinical datasets. |
| MONAI | PyTorch-based deep learning framework for healthcare imaging. | Model Zoo, MONAI Bundle format, tutorials, technical documentation. | N/A |
| Mayo-UGA RadOnc Foundation Models | Models focused on advancing radiation oncology using AI. | NLP Database, Segment Anything Model (SAM), Radonc-GPT. | N/A |
| RadGenome-Chest CT Dataset | Large-scale region-guided 3D chest CT interpretation dataset. | 197 organ categories, 665K grounded reports, 1.3M VQA pairs. | CT-RATE |
| PMC-LLaMA | Open-source medical language model designed for multilingual medical QA tasks. | Surpasses GPT on medical QA benchmarks. | Medical corpus. |
| IU-Xray-Report-Generation | Project generating medical reports from chest X-ray images. | Hybrid architecture using CNNs and RNNs for image feature extraction and report generation. | IU X-ray dataset. |
Development of RadFound 🔗
RadFound is an advanced vision-language (VL) foundation model specifically designed for radiology. It is trained on an extensive dataset comprising over 8.1 million images and 250,000 image-text pairs, covering 19 major organ systems and 10 imaging modalities. This model introduces an enhanced vision encoder to capture intra-image local features and inter-image contextual information, alongside a unified cross-modal learning design tailored to radiology. RadFound has been benchmarked using RadVLBench, which includes tasks such as medical vision-language question-answering and text generation tasks like captioning and report generation. When tested on real-world benchmarks involving chest X-rays, mammograms, and thyroid CT scans, RadFound significantly outperformed other VL foundation models. (source)
RadFM: A Multimodal Approach 🔗
RadFM is another initiative aimed at developing a comprehensive radiology foundation model. This model leverages a large-scale Medical Multi-modal Dataset (MedMD), which includes 16 million 2D and 3D medical scans. RadFM employs an architecture that enables visually conditioned generative pre-training, allowing for the integration of text input with 2D or 3D medical scans to generate responses for diverse radiologic tasks. The model was initially pre-trained on MedMD and later fine-tuned on RadMD, a radiologic-specific dataset containing 3 million visual-language pairs. (source)
Medical AI Research Foundations 🔗
The Medical AI Research Foundations repository on PhysioNet is a valuable resource for open-source medical foundation models. It provides non-diagnostic models, APIs, and resources like code and data to accelerate medical AI research. The repository hosts models for chest X-ray and pathology, trained on publicly available clinical datasets. The models are designed to generate representations of medical images without producing diagnostic outputs. This initiative aims to democratize access to foundational medical AI models and facilitate rapid development of new solutions.
MONAI: A PyTorch-Based Framework 🔗
MONAI is an open-source framework for deep learning in healthcare imaging, built on PyTorch. It is part of the PyTorch Ecosystem and aims to provide a comprehensive toolkit for healthcare imaging. MONAI offers a Model Zoo for sharing the latest models from the community, and it supports workflows through the MONAI Bundle format. The framework includes technical documentation, examples, and notebook tutorials to help researchers and developers get started with building healthcare imaging models.
Mayo-UGA RadOnc Foundation Models 🔗
The Mayo-UGA RadOnc Foundation Models GitHub organization focuses on advancing Radiation Oncology through the application of Large Language Models (LLMs), Machine Learning Models (LMMs), and Artificial Intelligence (AI). The organization develops, evaluates, and applies cutting-edge technologies to improve patient care in Radiation Oncology and Radiology. Projects include a Radiation Oncology NLP Database, a Segment Anything Model (SAM) for Radiation Oncology, and Radonc-gpt, a large language model for radiation oncology. The organization encourages contributions from researchers, clinicians, and developers interested in advancing AI in radiology and radiation oncology.
RadGenome-Chest CT Dataset 🔗
The RadGenome-Chest CT dataset is a comprehensive, large-scale, region-guided 3D chest CT interpretation dataset based on CT-RATE. It includes organ-level segmentation for 197 categories, 665K multi-granularity grounded reports, and 1.3M grounded VQA pairs. This dataset is designed to enhance visual representation learning for computational pathology by exploiting large-scale image-text pairs gathered from public resources, along with domain-specific knowledge in pathology.
Knowledge-Enhanced Vision-Language Pre-Training 🔗
A knowledge-enhanced vision-language pre-training approach has been proposed for auto-diagnosis on chest X-ray images. This approach involves training a knowledge encoder based on an existing medical knowledge graph, which is then used to guide visual representation learning. The pre-trained knowledge encoder enhances the model's ability to perform zero-shot diagnosis of unseen brain diseases. This method is part of a broader effort to develop Radiology Foundation Models, such as RadFM, which aim to integrate text input with medical scans for diverse radiologic tasks. (source)
GPT-4V Evaluation 🔗
In a study evaluating the GPT-4V model, 92 radiographic cases, 20 pathology cases, and 16 location cases across 17 medical systems and 8 imaging modalities were assessed. The evaluation revealed that while GPT-4V shows promise, it is still far from clinical usage. This underscores the need for further research and development in the field of medical AI to achieve clinically viable solutions. (source)
PMC-LLaMA: An Open-Source Language Model 🔗
PMC-LLaMA is an open-source language model acquired by leveraging a large medical corpus. It surpasses chatGPT on medicalQA benchmarks, demonstrating its potential for medical question-answering tasks. The model is part of a broader effort to develop multilingual language models for medicine, benefiting a linguistically diverse audience from different regions. This initiative aims to enhance the accessibility and applicability of medical AI across various languages and cultures. (source)
Conclusion 🔗
The development of radiology foundation models on GitHub is a rapidly evolving field, with numerous initiatives aimed at enhancing the capabilities of AI in medical imaging. From RadFound's expert-level vision-language model to RadFM's multimodal approach, these projects are pushing the boundaries of what is possible in radiology and healthcare imaging. Open-source repositories like Medical AI Research Foundations and MONAI provide valuable resources for researchers and developers, while initiatives like Mayo-UGA RadOnc Foundation Models and RadGenome-Chest CT dataset contribute to the advancement of AI in radiation oncology and computational pathology. As these efforts continue to evolve, they hold the potential to significantly improve patient care and clinical outcomes in the field of radiology.
Key Datasets and Architectures in Radiology Foundation Models 🔗
| Dataset Name | Description | Size/Scope | Usage/Application |
|---|---|---|---|
| MedMD | A large-scale multi-modal medical dataset containing 2D and 3D medical scans. | 16 million 2D/3D medical scans | Training multimodal radiology models like RadFM through visually conditioned pre-training. |
| RadMD | A filtered, radiology-specific subset of MedMD. | 3 million visual-language pairs | Fine-tuning RadFM for radiology-specific tasks using supervised visual instruction tuning. |
| RadVLBench | Benchmark dataset for evaluating vision-language models in radiology. | 8.1 million images, 250,000 image-text pairs | Evaluation of models like RadFound on vision-language tasks such as question-answering and report generation. |
| IU X-ray Dataset | Dataset containing chest X-ray images paired with medical reports. | 7,470 images with reports | Used for report generation tasks by models like IU-Xray-Report-Generation. |
| RadGenome-Chest CT Dataset | A large-scale, region-guided 3D chest CT interpretation dataset. | 197 organ categories, 665K grounded reports, 1.3M VQA pairs | Enhancing visual representation learning in computational pathology and radiology. |
| CT-RATE | Dataset providing ground truth for chest CT segmentation tasks. | Large-scale, organ-level segmentation | Supports the RadGenome-Chest CT dataset for region-based 3D chest CT interpretations. |
| PMC-LLaMA Medical Corpus | Large medical corpus used for training multilingual medical language models. | Extensive text corpus in the medical domain | Used for developing and fine-tuning language models like PMC-LLaMA for medical QA tasks. |
| AbdomenAtlas Dataset | Dataset of CT volumes with voxel-wise annotations of organs and pseudo annotations for tumors. | 9,262 CT volumes, 25 organs, 7 tumor types | Used for supervised pre-training of models for improved efficiency in medical tasks. |
| RadBench | Evaluation benchmark for radiology foundation models. | Multiple tasks including diagnosis, question-answering | Benchmarking models like RadFM on tasks like disease diagnosis, report generation, and visual question answering. |
Large-Scale Medical Datasets 🔗
In the realm of radiology foundation models, the availability and utilization of large-scale datasets are paramount. One notable dataset is the MedMD, which comprises 16 million 2D and 3D medical scans. This dataset is pivotal for training models like RadFM, enabling them to handle diverse radiologic tasks through visually conditioned generative pre-training. MedMD stands out as it includes both 2D and 3D scans, making it one of the first multi-modal datasets to offer such a comprehensive scope.
Another significant dataset is the RadMD, a filtered version of MedMD, specifically curated for radiology. It contains 3 million radiologic visual-language pairs, providing a domain-specific dataset for fine-tuning models. This dataset spans various data formats and modalities, enhancing the ability of models to perform supervised visual instruction-tuning.
In addition to these, the IU X-ray dataset offers chest X-ray images paired with medical reports. This dataset is crucial for projects aimed at generating medical reports from X-ray images, leveraging deep learning techniques to interpret images and produce accurate reports.
Vision-Language Model Architectures 🔗
The development of vision-language models in radiology has seen significant advancements, with architectures designed to integrate visual and textual data seamlessly. The RadFound model introduces an enhanced vision encoder that captures both intra-image local features and inter-image contextual information. This model employs a unified cross-modal learning design tailored to radiology, allowing it to perform tasks such as medical vision-language question-answering and report generation.
Similarly, RadFM employs a unique architecture that enables visually conditioned generative pre-training. This approach allows for the integration of text input with 2D or 3D medical scans, facilitating diverse radiologic tasks. The architecture is designed to handle multi-image input and visual-language interleaving, making it highly adaptable to practical clinical scenarios.
Benchmarking and Evaluation Frameworks 🔗
Benchmarking and evaluation frameworks are critical for assessing the performance of radiology foundation models. The RadVLBench is a benchmark specifically constructed to evaluate models like RadFound. It includes tasks such as medical vision-language question-answering and text generation, providing a comprehensive assessment of the model's capabilities.
Similarly, the RadBench framework is designed to evaluate RadFM. It comprises five tasks: modality recognition, disease diagnosis, visual question answering, report generation, and rationale diagnosis. RadBench allows for both automatic and human evaluation, ensuring a thorough assessment of the model's ability to handle practical clinical problems.
Open-Source Code Repositories 🔗
Open-source code repositories play a crucial role in the development and dissemination of radiology foundation models. The RadFM GitHub repository provides the official code for the RadFM model, along with links to the MedMD dataset and model checkpoints. This repository facilitates collaboration and further research by making all data, codes, and models publicly available.
Another important repository is the IU-Xray-Report-Generation project, which focuses on generating medical reports from chest X-ray images. It uses a hybrid architecture combining convolutional neural networks (CNNs) for image feature extraction and recurrent neural networks (RNNs) for report generation. The repository includes the necessary code and dataset links, encouraging contributions to improve the project.
Integration of AI in Clinical Practice 🔗
The integration of AI in clinical practice is a growing trend, with radiology foundation models playing a pivotal role. Models like RadFound and RadFM are designed to be integrated into clinical workflows, offering solutions for tasks such as disease diagnosis and report generation. The ability of these models to process multimodal data—combining images and text—makes them highly suitable for real-world applications.
The Mayo-UGA RadOnc Foundation Models organization is another example of AI integration in clinical practice. It focuses on advancing radiation oncology through the application of large language models (LLMs) and machine learning models (LMMs). The organization encourages contributions from researchers and clinicians, fostering collaboration to improve patient care in radiation oncology and radiology.
In summary, the development of radiology foundation models is heavily reliant on large-scale datasets, advanced architectures, and comprehensive evaluation frameworks. Open-source repositories and the integration of AI in clinical practice further enhance the potential of these models to revolutionize the field of radiology.
Evaluation and Benchmarking of Radiology Foundation Models 🔗
Evaluation Metrics and Methodologies 🔗
In the realm of radiology foundation models, evaluation metrics and methodologies are crucial for assessing model performance. The RadBench framework is a comprehensive evaluation tool designed to assess radiology models' capabilities. It includes tasks such as modality recognition, disease diagnosis, visual question answering, report generation, and rationale diagnosis. This framework employs both automatic and human evaluation methods, ensuring a thorough assessment of the model's ability to handle practical clinical problems. The automatic evaluation involves quantitative metrics, while human evaluation focuses on clinical relevance and accuracy.
Another notable evaluation benchmark is the RadVLBench, which is specifically constructed to evaluate models like RadFound. It includes tasks such as medical vision-language question-answering and text generation, providing a comprehensive assessment of the model's capabilities. These benchmarks are essential for comparing different models and identifying areas for improvement.
Comparative Analysis of Foundation Models 🔗
The comparative analysis of foundation models is a critical aspect of evaluating their performance. The RadFM model has been benchmarked against state-of-the-art (SOTA) models like OpenFlamingo and MedVInT. The results indicate that RadFM significantly outperforms existing multi-modal foundation models. This superiority is evident in various tasks, including modality recognition, disease diagnosis, and visual question answering. The evaluation involves a radar plot that visualizes the average performance across different metrics, highlighting RadFM's strengths.
Moreover, the RadMD dataset plays a crucial role in fine-tuning RadFM for domain-specific tasks. This dataset contains 3 million radiologic visual-language pairs, providing a rich resource for supervised visual instruction-tuning. The comparative analysis of RadFM with other models demonstrates its ability to handle diverse radiologic tasks effectively.
Benchmarking on Diverse Datasets 🔗
Benchmarking foundation models on diverse datasets is essential for evaluating their generalizability and robustness. The MedMD dataset is a large-scale medical multi-modal dataset comprising 16 million 2D and 3D medical scans. This dataset is pivotal for training models like RadFM, enabling them to handle diverse radiologic tasks through visually conditioned generative pre-training. MedMD stands out as it includes both 2D and 3D scans, making it one of the first multi-modal datasets to offer such a comprehensive scope.
Additionally, the AbdomenAtlas dataset is an extensive dataset of 9,262 CT volumes with per-voxel annotation of 25 organs and pseudo annotations for seven types of tumors. This dataset enables the supervised pre-training of AI models at scale, enhancing their performance and efficiency compared to self-supervised pre-training. The benchmarking on these datasets provides insights into the models' ability to generalize across different clinical scenarios.
Human and Automated Evaluation Techniques 🔗
Human and automated evaluation techniques are integral to the assessment of radiology foundation models. The GREEN framework is a radiology report generation metric that leverages the natural language understanding of language models to identify and explain clinically significant errors in candidate reports. This framework employs both automated and human evaluation methods, ensuring a comprehensive assessment of the model's performance.
Automated evaluation involves quantitative metrics such as BLEU, METEOR, and ROUGE scores, which measure the accuracy and coherence of generated reports. Human evaluation, on the other hand, focuses on clinical relevance and accuracy, providing a qualitative assessment of the model's ability to generate clinically useful reports. The combination of these techniques ensures a robust evaluation of the models' capabilities.
Challenges and Future Directions in Evaluation 🔗
Despite the advancements in evaluation methodologies, several challenges remain in the assessment of radiology foundation models. One of the primary challenges is the lack of standardized evaluation metrics across different models and tasks. This inconsistency makes it difficult to compare models and identify areas for improvement. Moreover, the reliance on large-scale datasets for training and evaluation poses challenges in terms of data availability and quality.
Future directions in evaluation involve the development of standardized benchmarks and metrics that can be applied across different models and tasks. Additionally, there is a need for more comprehensive evaluation frameworks that consider both quantitative and qualitative aspects of model performance. The integration of human expertise in the evaluation process is also crucial for ensuring the clinical relevance and accuracy of the models.
In conclusion, the evaluation and benchmarking of radiology foundation models are critical for assessing their performance and identifying areas for improvement. The use of comprehensive evaluation frameworks, such as RadBench and RadVLBench, provides valuable insights into the models' capabilities. However, challenges remain in terms of standardization and data availability, highlighting the need for continued research and development in this field.
Conclusion 🔗
The research report highlights significant advancements in the development of radiology foundation models, particularly through initiatives like RadFound and RadFM. These models leverage extensive datasets, such as MedMD, to enhance their capabilities in handling diverse radiologic tasks through multimodal approaches. The introduction of comprehensive evaluation frameworks like RadBench and RadVLBench emphasizes the importance of rigorous benchmarking in assessing model performance, showcasing how these models significantly outperform existing solutions in various tasks.
The implications of these findings are profound for the future of medical imaging and AI integration in clinical practice. As open-source resources like the Medical AI Research Foundations and MONAI continue to democratize access to foundational models, they pave the way for enhanced collaboration among researchers and clinicians. Moving forward, addressing challenges related to standardization in evaluation metrics and ensuring the availability of high-quality datasets will be crucial for the continued evolution of these technologies. The ongoing development and refinement of radiology foundation models hold the potential to significantly improve patient care and clinical outcomes in the field of radiology.
References 🔗
- https://github.com/chaoyi-wu/RadFM
- https://chaoyi-wu.github.io/RadFM/
- https://github.com/ChantalMP/RaDialog
- https://github.com/openmedlab/MedFM
- https://github.com/MedAIerHHL/CVPR-MIA
- https://github.com/harrison-ai/radbench
- https://arxiv.org/abs/2308.02463
- https://github.com/Stanford-AIMI/green
- https://github.com/markin-wang/xpronet
- https://github.com/Mayo-Clinic-RadOnc-Foundation-Models/
- https://github.com/openlifescience-ai/Awesome-AI-LLMs-in-Radiology/blob/main/README.md
- https://github.com/MrGiovanni/SuPreM
- https://paperswithcode.com/paper/expert-level-vision-language-foundation-model