Data scarcity is a perennial problem when applying deep learning (DL) to medical imaging. In vision tasks related to natural images, DL practitioners often have access to astoundingly large annotated data sets on which they can train. However, due to privacy concerns and the expense of creating them, access to large annotated data sets is rare in medical imaging. The natural follow-up question is: How can practitioners in the field of medical imaging best use DL given limited data? In this article, I’ll discuss one approach to stretch the use of available data, called self-supervised learning.
Self-supervised learning is a form of unsupervised learning in which a computational model learns a desired task without human supervision. Human supervision can come in many forms, but, in this article, by “human supervision” we mean human-generated annotations of data. In computer vision, these annotations are: classes for classification tasks, masks for segmentation tasks, and target images for regression tasks. In general, these annotations are expensive to obtain; if we can bypass the requirement or reduce the burden of creating annotations, we could use DL methods in many more tasks.
A central idea in DL is automatically finding efficient representations that capture salient features of the data. This is in contrast to previous approaches where machine learning practitioners hand-crafted representations that they thought were meaningful to solving the problem (for example, finding edges, corners, and blobs in images). In DL there has been a push to move away from these hand-crafted representations. So much so that the word “hand-crafted” has become a pejorative. In contrast to hand-crafted representations, data-driven representations are claimed to be near optimal for almost any task (so long as there is sufficient data to automatically find these representations).
What is self-supervised learning exactly? 🔗
Self-supervised learning is a subset of unsupervised learning where representations are learned from signals inherent in the data instead of human-generated annotations. The method was popularized in the field of natural language processing (NLP) where researchers had huge amounts of unannotated data and wanted to learn a transferable representation. For example, NLP researchers trained a neural network to predict the next word in a sentence (a task that does not involve human-generated annotations). The researchers found that the intermediate representation was useful for a variety of NLP tasks. The success of self-supervised learning techniques in NLP spurred research in computer vision in the last five or so years (to the best of our knowledge) because of its success at representation learning.
One of the first papers to popularize self-supervised learning in computer vision is the 2015 ICCV paper by Doersch, Gupta, and Efros. The method proposed in the paper is to train a deep network to predict where an image patch is in relation to another image patch (for example, to the right, left and below, or above). The motivating idea is that a network that can accomplish this task on a variety of natural images will have learned about the structure of the underlying data (for example, “eyes are above noses” or “ground is below the sky”). The hypothesis behind learning this task is that in learning this semantic information about the data, an efficient representation will have been learned without the use of external annotations.
That exemplar illustrates the methodology behind the entire area of self-supervised learning, namely, training a network to learn a non-trivial relationship held within the data. An important consideration is that the utility of the found representation will be upper bounded by the task’s ability to learn meaningful information about the data; however, there is no reliable method for determining what task will generate good representations for a particular domain. Regardless, several good self-supervised learning tasks have been found to create good transferable representations for data without the use of human-generated annotations.
Why can’t I just use a network pre-trained on ImageNet? 🔗
A common way to create good representations for natural images is to use a network pre-trained on ImageNet–a popular data set of over 14 million images of 1000 classes (for example, “tiger cat” and “teapot”). To pre-train a network using ImageNet, you first train it to classify these millions of example images into the 1000 classes. Then, you train the network for a related task, under the assumption that the pre-training will improve performance; this methodology is an example of transfer learning.
The common consensus about using ImageNet pre-trained networks is that these networks have learned a huge amount about natural images because the diversity of the data and the difficulty of the task. The claim is that these networks have learned useful ways to process images; in fact, the argument goes, these pre-trained networks have learned ways to process images that correspond to human perception. Thus pre-trained networks should transfer broadly across tasks, especially within the domain of natural images.
In many cases, transfer learning with ImageNet pre-trained networks has been shown to be useful. The networks converge faster and to better solutions when data is limited. However, researchers have shown that ImageNet pre-training doesn’t help all that much when you have access to a moderate amount of data.
In contrast to natural images, the utility of ImageNet pre-trained networks on medical images has generally been low. The convolutional filters that are useful to process natural images do not necessarily transfer to medical images; natural images have specific statistical characteristics that differ from medical images. While researchers have shown that ImageNet pre-trained networks can aid network training in medical images, the inconsistent performance motivates the need for alternative methods that work more consistently; this is where self-supervised learning may help.
Self-supervised learning can serve the same function as ImageNet pre-training, but self-supervised learning can be more easily tailored to a new domain. One of the reasons that ImageNet is used for pre-training is because it is large and diverse. But annotations are expensive, so people do not want to collect a huge number of annotations for their specific domain; however, these annotations provide a strong signal to the network to learn discriminative features. Self-supervised learning proposes an alternative solution because it provides a strong signal via an annotation on a diverse difficult task; the trick is that the annotation is automatically generated from the data instead of a human.
How do I use self-supervised learning in medical images? 🔗
There have been a few papers using self-supervised learning techniques to learn useful representations for medical images; however, self-supervised learning with medical images appears less explored than with natural images. I’ll review a few methods below separated by the underlying task.
Determining spatial relationship 🔗
As previously mentioned, the Doersch method of self-supervised learning involves learning the spatial relationship between patches of an image. For example, if you have a patch containing the face of a human in an upright orientation and you have another patch containing the torso, the task would be to learn that the face is above the torso and that the torso is below the face.
A 2019 MICCAI paper by Blendowski, Nickisch, and Heinrich (preprint here) explores learning spatial relationships in 3D structural computed tomography (CT) images. They propose two models: 1) a 3D version of the Doersch method and 2) a method to predict how much another image patch is offset to another image. Below is a short description of the proposed methods; we also created two Jupyter notebooks which show example code for the first method and the second method, in which we train the methods on the IXI data set of head MR images. Hopefully these notebooks can make some of these ideas more concrete and serve as a reference for implementation.
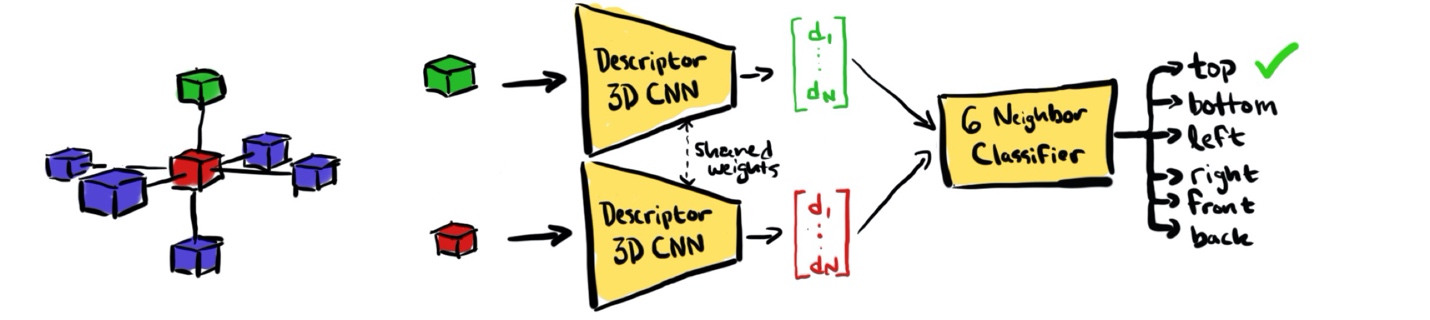
Figure 1 diagrams the architecture of the 3D Doersch-style method. The task is to take two cubic patches—one is the reference and the other is the query—and determine where in space the query patch is in relation to the reference (in Figure 1, the red cube is the reference patch and the green cube is an example query patch). The two patches are independently fed into a CNN which makes a reduced-dimension descriptor vector out of each patch, these two feature vectors are then concatenated and fed into an auxiliary neural network classifier that determines if the query patch is above, below, to the left, to the right, to the front, or to the back of the reference patch.
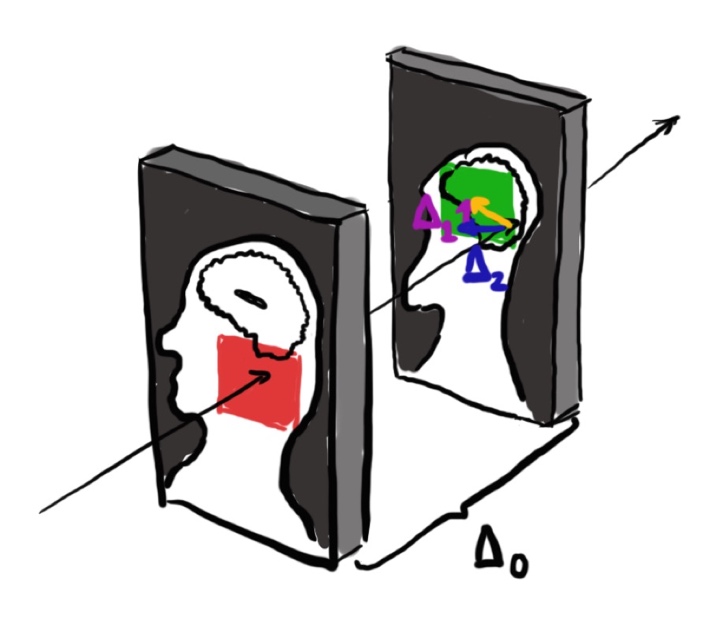
The second, proposed method is different; in this approach, the authors propose to take two patches and regress the offsets of the query patch to the reference patch. For an illustration of the method, see Figure 2 which shows the patch setup in a sagittal view of a head image where the red square represents the reference patch and the green square represents the query patch. To further explain, in this method the authors propose to use a pseudo-3D model (that is, a model that uses 2D convolutions but uses three adjacent slices as input to the network) and take two pseudo-3D patches that are \(\Delta_0\) units away from each other and offset the center of the query patch by two random quantities \(\Delta_1\) and \(\Delta_2\) which correspond to the in-plane image axes. The task is then to regress these two quantities \(\Delta_1\) and \(\Delta_2\).
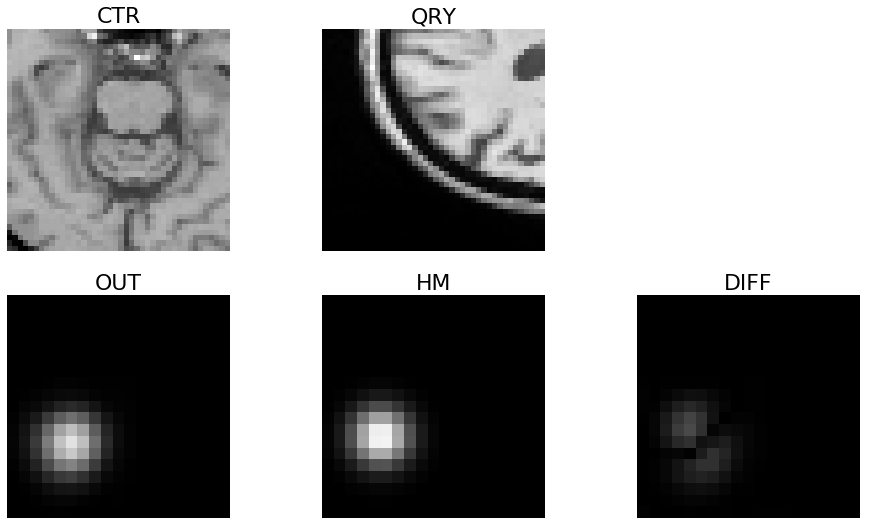
In practice, the authors find that directly regressing the values of \(\Delta_1\) and \(\Delta_2\) is sub-optimal. Consequently, they propose a modification to regress a heatmap that highlights the values of \(\Delta_1\) and \(\Delta_2\). The heatmap that they try to regress is a 2D image of an offset Gaussian kernel where the offset is determined according to \(\Delta_1\) and \(\Delta_2\). An example of such a heatmap is shown in Figure 3 under the title “HM.” To get an idea of how this particular heatmap was generated, we can look at the two image patches that determined the offsets. The reference image patch (titled “CTR”) and the query image patch (titled “QRY”) are offset from one another where the query image is down and to the left of the reference patch. As we can see, this offset relationship is captured in the heatmap; that is, the center of the Gaussian blob is down and to the left as well. This figure is from the Jupyter notebook we mentioned previously (link) where we trained a network to do this task, and it predicted a heatmap shown in Figure 3 (titled “OUT”) which closely resembles the true heatmap image. This result shows that the network has learned how an image is offset to another image in a certain plane. (Note that in Figure 3 the image titled “DIFF” shows the absolute difference between “OUT” and “HM”.)
To tie the above methods back to representation learning, it is important to note that getting the network to perform optimally on the above tasks is not of primary concern; performing well on the spatial relationship task is not important outside of the transferability of the trained network to some other task (for instance, image segmentation). Ultimately, the authors of the above paper found that a network pre-trained with the heatmap method out-performed a network pre-trained on the 3D Doersch method on a segmentation task!
In-painting 🔗
Another interesting and easy-to-implement task for self-supervised learning is that of in-painting. The idea is developed in the 2016 CVPR paper where Pathak and others introduced the idea of context encoders which are networks that are trained to fill in missing portions of images. This task falls into self-supervised learning because, given a normal image, we can remove portions of that image to simulate missing data. Then the image with simulated missing data is input to a deep network and the unmodified image as the target, and the network is trained with some loss function like L1 or MSE.
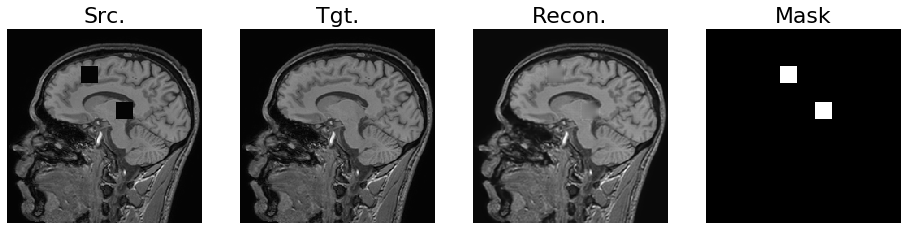
For reference, we created an example Jupyter notebook showing an application of context encoders for medical images (again using the IXI data set of head MR images). In Figure 4, we show an example image with missing portions (titled “Src.”) and the resulting in-painted image (titled “Recon.”). The true image without missing data is also shown in Figure 4 under the title “Tgt.”
The motivating idea of the in-painting task is that the network will have to learn how to fill in the missing portion of the image from contextual cues around the missing portion. If the network has the ability to fill in missing data based on contextual cues, it will—the argument goes—have learned a significant amount of semantic information about the data.
Note that Pathak and others found that simply regressing the missing data with a loss function like MSE or L1 loss was not sufficient. They needed to use a discriminator network in addition to the regression loss term to better enforce realistic filled-in images; this combination resulted in better transferability to down-stream tasks.
To the best of our knowledge, context encoders have not been extensively used in medical images, although a journal article using a similar method can be found here. Another use case for context encoders in medical images has been for anomaly detection.
Reconstructions, rotations, and other tasks 🔗
There are two other tasks which we should highlight: reconstruction and rotation prediction. A reconstruction task is simply taking an input image and compressing to a reduced-size representation and using that reduced-sized representation to reconstruct the original input image. This is the idea behind auto-encoders which are a familiar topic to most DL practitioners. This idea has a long history of being used to pre-train deep neural networks; however, autoencoders are often difficult to make useful—they can learn a degenerate function that isn’t transferable. And, in general, you cannot use them with certain architectures that are common in medical images; for instance, the U-Net because that network will use the skip connections to pass through the original image to the output and, consequently, not learn anything about the data.
Another method which has shown a good amount of success in several domains is to predict how an image is rotated. The motivating idea is that images often have a consistent ordering given an orientation; for example, the eyes are above the nose in an image of an upright person. So, if you learn how to predict a rotation, you will have learned the normal order of objects or structures in an image. The reconstruction and rotation prediction tasks in medical imaging are explored here.
For completion’s sake, another example of self-supervised learning is in the use of longitudinal images to learn if an image comes from the same patient or a different patient. The longitudinal aspect of this work makes this less broadly applicable, but, if you have access to data with annotations like timestamps, using this information for a self-supervised learning approach—as the authors did here—could aid in down-stream applications.
Why isn’t self-supervised learning used more? 🔗
Given the claim that using a self-supervised pre-trained network results in a substantial improvement in performance, why isn’t self-supervised learning used more often? We speculate it is because: 1) it is difficult to come up with self-supervised tasks on which to pre-train a network and 2) it is a time-consuming to develop a DL method that effectively uses the representations provided by the self-supervised network.
Now that the seeds of self-supervised learning have been planted for medical imaging with the previously described papers, we may see more DL practitioners using self-supervised learning to improve the performance of their deep networks.
Takeaways 🔗
Self-supervised learning is an unsupervised learning 1 approach to representation learning. The approach makes use of data without annotations to help learn down-stream tasks, like image segmentation. Since unannotated data is easier to obtain than annotated data, we can make better use of all of the data we have access to by using self-supervised learning to pre-train a network on a task like the ones discussed above. Then we can use the resultant pre-trained network for fine-tuning on another task or in some other machine learning pipeline.
While the self-supervised learning tasks described above may not work well with every down-stream application, the underlying idea of using information inherent in the data to train a deep neural network is broadly useful, and it can be applied to stretch the use of available data in medical imaging.
Contributions 🔗
Jacob was the primary author, David provided editorial feedback, and Yujan provided substantive feedback and the topic for the review.
Footnotes 🔗
-
The definition of “Unsupervised learning” may require the input data to have “no pre-existing labels” in some contexts (e.g., K-means clustering) but “minimum human supervision” in others (e.g., autoencoders). We will use the latter requirement in this article. In either case, the end goal is transferable representation learning. ↩

