An overview of neural net architectures beyond the U-Net 🔗
Innovation in neural networks often happens through architectural change. In fact, architectural change is arguably what popularized deep neural networks (DNNs). The convolutional variety of DNNs had their first mainstream success in 1989 for classification in a now-antiquated model called the LeNet. In 2012, the AlexNet was introduced for a more difficult dataset, but the blocks which composed the AlexNet were mostly the same as the blocks which composed the LeNet. The innovation of the AlexNet was mostly in how the blocks were situated with respect to one another and the number of blocks used—in other words, the architecture1.
If architecture plays such an important role in performance, it is natural to ask how to choose an architecture. In computer vision there have been a slew of papers that introduce both radically different architectures and incremental improvements. In medical image segmentation, however, the architecture often seems to default to the U-Net.
The U-Net is a simple-to-implement DNN architecture that has been wildly successful in medical imaging; the paper that introduces the U-Net, published in 2015, is the most cited paper at the prestigious medical imaging conference MICCAI. Over the past five years, however, there have been significant improvements to architectures for segmentation tasks, and it is no longer safe to assume that the U-Net will provide near state-of-the-art performance.
This article will provide an overview of the U-Net, explaining why it works and why it is the default, and then it discusses a few more recent architectures which are promising for medical imaging applications.
What is the U-Net and why is it so popular? 🔗
The U-Net is a 2D or 3D fully-convolutional neural network architecture that consists of an encoder and a decoder with skip connections that connect the two at various levels of feature abstraction.
The encoder consists of convolutional layers along with progressive downsampling which captures contextual information about the input in a compressed representation. The decoder consists of convolutional layers with progressive upsampling which, along with the skip connections, preserves sharp boundaries and structural information about the input image. Figure 1 shows an example of a U-Net architecture.

The purpose of downsampling—in this case via max-pooling—in the decoder is not only to increase the receptive field2 of the network, but make it computationally and memory efficient to create a large number of convolutional feature maps. More feature maps means the network can learn more complex relationships between the input and output.
The purpose of upsampling is to restore the spatial structure in the output so the prediction corresponds pixel- or voxel-wise to the input. This correspondence wouldn’t otherwise be the case due to downsampling. The skip connections restore the spatial structure at every level.
The downsampling and upsampling encoder-decoder structure combines to make the U-Net have a large receptive field while maintaining spatial structure in the prediction.
The U-Net is ubiquitous in medical imaging conferences and journal articles. The apparent reason is that it is established to work reasonably well and the model is easy to implement. Consequently it is a go-to for prototyping. As opposed to some of the other models that I will discuss, the U-Net can be written in PyTorch in a few lines, which makes it quick to prop up and debug.
For an example implementation, see here. I tried to keep the implementation short without doing anything outright barbarous. For intuition as to why this network is so often used, that example implementation took me about 30 minutes from scratch (although I’ve implemented this several times before).
What are the alternatives? 🔗
What options does a deep learning practitioner in medical imaging have outside of the U-Net? We’ll start from the beginning with the ResNet, a now-classic architecture, and move on to more recent architectures that build on the encoder-decoder architecture of the U-Net.
Alternative 1: ResNet 🔗
The ResNet architecture introduced residual connections—similar to the skip connections of the U-Net—which made it possible to train architectures with many more convolutional layers. The residual connections added the features of earlier layers to later ones so that the convolutional layers only had to learn the difference (that is, the residual). This can be seen in the equation below, where x is the input, y is the output, and f is the residual layer.
\[y = f(x) + x \implies f(x) = y - x\]This trick helped solve a long-standing problem of getting backpropagation to update the weights of early layers in very deep neural networks.
The ResNet was initially proposed for classification tasks, and thus was a straight-forward architecture composed of residual blocks and pooling layers to downsample the input to an output vector shaped according to the number of target classes. The downsampling and lack of upsampling layers make this architecture not immediately transferable to segmentation, but the architecture can be modified to do so.
Another problem with this network for medical imaging purposes is that the implementations of the network are for 2D networks. When working with 3D images like MR and CT, you’ll either need to use a 2D or 2.5D method of slice extraction to fit the images into the network. Given that the architectures that we will discuss in the remainder of this article are created for natural images, this 2D dimension problem persists; however, all of the mentioned 2D networks can be converted into 3D networks by replacing 2D convolutions, pooling, and other layers for their 3D equivalents. This conversion can make the number of parameters explode, and the resulting network can be infeasible to train due to memory-constraints. Nevertheless, it is usually straight-forward to modify the hyperparameters so as to create a working 3D model from a 2D one.
Alternative 2: DeepLabv3+ 🔗
The DeepLabv3+ architecture is one way to modify the ResNet for semantic segmentation. There are several modifications, but the one most relevant to our purposes is the modification to be an encoder-decoder with skip connections (akin to the U-Net). This is done by making the ResNet into the encoder and adding additional upsampling layers to form the decoder. Skip connections use features from intermediate layers of the ResNet and concatenate them to intermediate features in the decoder. See Figure 2 for a block diagram of the architecture (look at the original paper for a description of the ASPP block).

Another important modification is that the architecture uses dilated convolutions instead of downsampling to increase the receptive field. See Figure 3 for an example of a dilated convolution, where the shaded blue squares in the array on the bottom show the kernel of the dilated convolution (mapping to the shaded teal square on the top array).
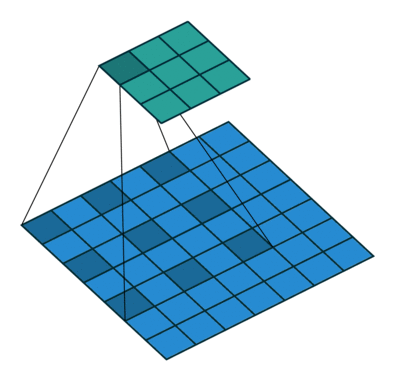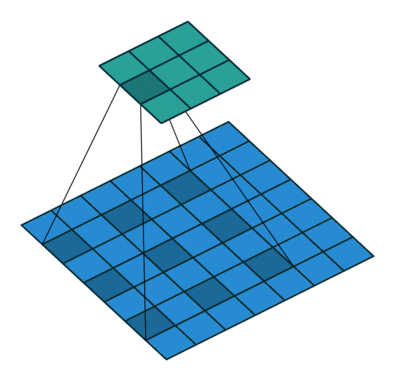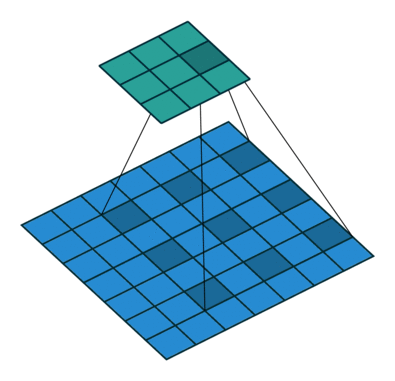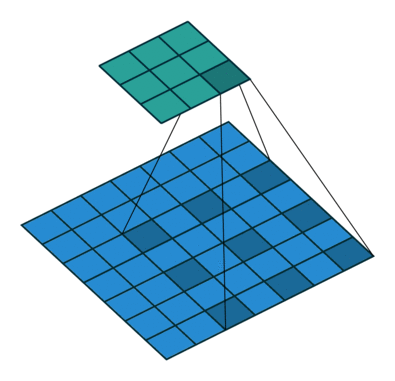
The benefit of dilated convolution is the size of the feature map is kept constant while increasing the receptive field. While keeping the feature maps a constant size is more computationally and memory inefficient, the resulting dense features make it easier to resolve small objects in the input image—an important characteristic for segmentation.
These two modifications have been shown to produce state-of-the-art performance on many semantic segmentation datasets in medical imaging. While the network is large in terms of numbers of parameters and computationally expensive to run, this architecture has the benefit of using a ResNet as the encoder which can be found pre-trained in all of the most common deep learning frameworks (for example, PyTorch and Tensorflow). Using a pre-trained network can often reduce the training time or improve the robustness of the network on an underlying task (as discussed in our previous article on self-supervised learning).
There are many implementations of this network that are freely available, but many of the PyTorch implementations we found were difficult to follow, so we implemented our own that attempts to maximize code reuse from the PyTorch vision repository.
Alternative 3: Tiramisu 🔗
The main contribution of the ResNet—the use of residual connections—begged the question: If it is good to connect previous layers output to later layers, is it better to connect all previous layers to all future ones? It turns out, in some applications, that the answer was yes. The DenseNet, composed of several densely-connected (in terms of residual connections) blocks, improved the performance on image classification tasks. An example of a densely-connected block is shown in Figure 4. Like the ResNet, however, DenseNets were designed for classification and need to be modified for semantic segmentation.

The Tiramisu architecture blends together the DenseNet and the encoder-decoder with skip connections structure of the U-Net to get the benefits of the densely-connected convolutional layers, as well as the greater spatial-structure-preserving aspects of the U-Net.
The idea of how to blend the two architectures together is simple, but it works well for medical image analysis applications. The idea is simply to replace the convolutional blocks of the U-Net with densely-connected blocks. If you look at the block diagram of the architecture in Figure 5, the resemblance is clear.

The use of densely-connected blocks makes this architecture more difficult to implement and debug, and makes it much less memory-efficient because it requires the storage of many intermediate features for later concatenation. Regardless, the use of densely-connected layers reduces the number of required parameters and can achieve state-of-the-art performance in difficult, small-dataset semantic segmentation tasks like multiple sclerosis lesion segmentation.
We provide an implementation of this network in both 2D and 3D here.
Alternative 4: Attention 🔗
Another recent modification to DNN architectures is the use of attention blocks; for example, Attention Gated Networks, ResNest, and Squeeze-excite networks. There are two main types of attention in convolutional networks: spatial and channel attention.
Spatial attention allows for long-range dependencies which allow the network to relate relatively distant objects in the image; without spatial attention, convolutional layers are restricted to calculating local features corresponding to the size of the convolutional kernel. An alternative formulation of spatial attention is to “focus” the network on certain regions of the image while ignoring other regions.
Channel attention computes what amounts to an importance weighting scheme of the channels (that is, feature maps) for the output of a convolutional layer. The weighting scheme attenuates certain feature maps so as to focus the “attention” of the network on the un-attenuated channels.
Depending on the task and the formulation of attention, adding attention blocks to your network architecture may help improve performance. For what it’s worth, I have had limited success employing either spatial or channel attention. Spatial attention, as formulated here, is a memory-hog and didn’t significantly improve performance on a variety of tasks. Likewise for channel attention. Attention gated networks seems to be a formulation of spatial attention that is less memory-intensive and shows promising results on medical image segmentation tasks, but the authors state that training is more complicated than would be otherwise. We implemented a version of the attention gates described in that architecture here.
Takeaways 🔗
We reviewed several neural network architectures, describing how they may or may not improve on the U-Net. We also provided some implementations which can be used to test out the described networks without too much pain. Ultimately, to get the most performance out of a DNN in any given task, you must try several architectures and pick the one that happens to works best. While architecture choice is important, choosing the right loss function and setting the right hyperparameters is also vital to squeezing the most performance out of the DNN.
You don’t need to go back to the LeNet and test every conceived architecture; you can take the DeepLabv3+ and the Tiramisu model and do hyperparameter sweeps on both to determine which works best for your application. In the future, innovation in architectures will provide significant benefits over the Deeplabv3+ and the Tiramisu, much like these models improve on the U-Net, and there’ll be new models to test that just work better across the board. However, there will never be a silver bullet for choosing an architecture and training a DNN, and the optimal choice will continue to evolve with the field of machine learning.

